Abstract
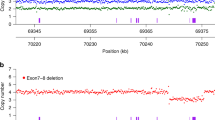
Spinal muscular atrophy (SMA) is the second most common lethal, autosomal recessive disease in Caucasians (after cystic fibrosis). Childhood SMAs are divided into three groups (type I, II and III), which are allelic variants of the same locus in a region of ∼850 kb in chromosome 5q12–q13, containing multiple copies of a novel, chromosome 5–specific repeat as well as many atypical pseudogenes. This has hampered the identification of candidate genes. We have identified several coding sequences unique to the SMA region. A genomic fragment detected by one cDNA is homozygously deleted in 17/29 (58%) of type I SMA patients. Of 235 unaffected individuals examined, only two showed the deletion and both are carriers of SMA. Our results suggest that deletion of at least part of this novel gene is directly related to the phenotype of SMA.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Dubowitz, V. Muscle disorders of childhood.(Saundere, Philadelphia, 1978).
Pearn, J. H. Infantile motor neuron diseases, in Advances in Neurology(ed. Rowland, L. P. ) 121–130 (Raven Press, New York, 1982).
Pearn, J. Classification of spinal muscular atrophies. Lancet 1, 919–922 (1980).
Brunstowicz, L. et. al Genetic mapping of chronic childhood–onset spinal muscular atrophy tochromosome 5q11. 2–13. 3.Nature 344, 540–541 (1990).
Gilliam, T.C. et al. Genetic homogeneity between acute and chronic forms of spinal muscular atrophy. Nature 345, 823–825 (1990).
Melki, J. et al. Gene for chronic proximal spinal muscular atrophies maps to chromosome 5q. Nature 344, 767–770 (1990).
Melki, J. et al. Mapping of acute (type I) spinal muscular atrophy to chromosome 5q12–q14. Lancet 336, 271–273 (1990).
Daniels, R.J. et al. Linkage analysis of spinal muscular atrophy. Genomics 12, 335–339 (1992).
Simard, L.R. et al. Linkage study of chronic childhood-onset spinal muscular atrophy (SMA): confirmation of close linkage to D5S39 in French Canadian families. Genomics 14, 188–190 (1992).
Francis, M.J. et al. A contig of non-chimeric YACs containing the spinal muscular atrophy gene in 5q13. Hum. molec. Genet. 2, 1161–1167 (1993).
Wirth, B. et al. Large linkage analysis in 100 families with autosomal recessive spinal muscular atrophy (SMA) and 11 CEPH-families using 15 polymorphic loci in the region 5q11.2-q13. 3 Genomics 20, 84–93 (1994a).
DlDonato, C. J. et al. Association between Ag1–CA alleles and severity of childhood–onset spinal muscular atrophy (SMA). Am. J. hum. Genet. 56, 1218–1229 (1994).
Simard, L.R. et al. Linkage disequilibrium analysis of childhood–onset spinal muscular atrophy (SMA) in the French–Canadian population. Hum.molec. Genet. 3, 459–463 (1994).
Burghes, A.H.M. etel. A multicopy dinucleotide marker that maps close to the spinal muscular atrophy gene. Genomics 21, 394–402 (1994).
Thompson, T.G. et al. High resolution physical map of the region surrounding the spinal muscular atrophy gene. Hum. molec. Genet. 2, 1169–1176 (1993).
Sargent, C.A., Chalmers, J., Leversha, M., Affarar, N.A. & Ferguson-Smith, M.A. Identification of a novel repeat element mapping to 4loci on chromosome 5. Cytogenet. Cell Genet. 58, 1901 (1991).
Shipley, M. et al. Mutational analysis of a patient with mucopolysaccharidosis type VII, and identification of pseudogenes. Am. J. hum. Genet. 52, 517–526 (1993).
Danesino, C., Gimelli, G., Cuoco, C. & Ciccone, M.O. Triplex gene dosage effect of β–glucuronidase and possible assignment to band q22 in a partial duplication 7q. Hum. Genet. 58, 371–373 (1981).
Arrendondo-Vega, F.X., Arfin, S.M. & Smith, M. Regional assignment of human beta–glucuronidase on chromosome 7. Cytogenet. Cell Genet 37, 403–404 (1984).
Breton, C., Schorp, M. & Nahlon, J. L Isolation and characterization of the human–concentrating hormone gene and a variant gene. Molec. Brain. Res. 18, 297–310 (1993).
Predeutour, F., Szpirer, C. & Nahon, J.L. Assignment of the human promelanin-concentrating hormone gene (PMCH) to chromosome 12q23–q24 and two variant genes (PMCHL1 and PMCHL2) to chromosome 5p14 and 5q12–q13. Genomics 19, 31–37 (1994).
Selig, S. et al. The spinal muscular atrophy region contains novel expressed pseudogenes originating from a neuronal cadherin gene. EMBO J. (in the press).
Buckler, A.J. et al. Exon amplification: a strategy to isolate mammalian genes based on RNA splicing. Proc. natn. Acad. Sci. U.S.A. 88, 4005–4009 (1991).
Warringtion, J.A. et al. A radiation hybrid map of 18 growth factor, growth receptor, hormone receptor or neurotransmitter receptor genes on the distal region of the long arm of chromosome 5. Genomics 13, 803–808 (1992).
Sartor, H., Ehlert, F., Grzeschik, K.H., Muller, R. & Adolph, S. Assignment of two human cell cycle genes, CDC25C and CCNB1, to 5q31 and 5q12, respectively. Genomics 13, 911–912 (1992).
Pines, J. & Hunter, T. Isolation of a human cyclin cDNA: evidence for cyclin nRNA and protein regulation in the cell cycle and for interaction with p34cdc2. Cell 58, 833–846 (1989).
Takahasi, M., & Adelstein, R.S. Evidence for inserted sequences in the head region of nonmuscle myosin specific to the nervous system. J. biol. Chem. 267, 17864–17871 (1992).
Gilliam, T.C. et al. Deletion mapping of DNA markers to a region of chromosome 5 that cosegregates with schizophrenia. Genomics 5, 940–944 (1989).
Melki, J. et al. De novo and inherited deletions of the 5q13 region in spinal muscular atrophies. Science 264, 1474–1477 (1994).
Wirth, B. et al. Defining new boarders of the spinal muscular atrophy (SMA) candidate region by two new microsatellites and Isolation of cDNAs. Am. J. hum. Genet 55, A207 (1994b).
Belt, K.T., Carroll, M.C. & Porter, R.R. The structural basis of the multiple forms of human complement component C4. Cell 36, 907–914 (1984).
Carroll, M.C., Palsdottir, A., Belt, K.T. & Porter, R.R. Deletion of complement C4 and steroid 21–hydroxylase genes in the HLA class III region. EMBO J. 4, 2547–2552 (1985).
Morel, Y. et al. Gene conversions and rearrangements cause discordance between inheritance of forms of 21–hydroxylase deficiency and HLA types. J. clin. endo. Met. 68, 592–599 (1989).
Bristow, J., Tee, M.K., Gitelman, S.E., Mellon, S.H. & Miller, W.L. Tenascin-X: a novel extracellular matrix protein encoded by the human XB gene overlapping P450c21B. J. Cell Biol. 122, 265–278 (1993).
Shen, L. et al Structure and genetics of the partially duplicated gene RP located immediately upstream of the complement C4A and the C4B genes in the HLA class lll region. Molecular cloning. exon–intron structure, composite retroposon, and breakpoint of gene duplication. J. biol. Chem. 269, 8466–8476 (1994).
Pang, S.Y. et al. Worldwide experience in newborn screening for classical congenital adrenal hyperplasia due to 21-hydroxylase deficiency. Pediatrics 81, 866–874 (1988).
Miller, W.L. & Morel, Y. The molecular genetics of 21–hydroxylase deficiency. A. Rev. Genet. 23, 371–393 (1989).
Morel, Y., Bristow, J., Gitelman, S.E., Miller, W.L. Transcript encoded on the opposite strand of the human steroid 21–hydroxylase/complement C4 gene locus. Proc. natn. Acad Sci. U.S.A. 86, 6582–6586 (1989).
Klyen, P.W. et al. Construction of a yeast artificial chromosome contig spanning the spinal muscular atrophy disease gene region. Proc. natn. Acad. Sci. U.S.A. 90, 6801–6805 (1993).
Church, D.M. et al. Isolation of genes from complex sources of mammalian genomic DNA using exon amplification. Nature Genet. 6, 98–105 (1994).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Thompson, T., DiDonato, C., Simard, L. et al. A novel cDNA detects homozygous microdeletions in greater than 50% of type I spinal muscular atrophy patients. Nat Genet 9, 56–62 (1995). https://doi.org/10.1038/ng0195-56
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/ng0195-56