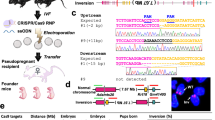
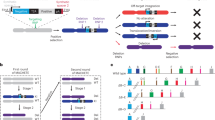
Abstract
Chromosomal deletions, especially nested deletions, are major genetic tools in diploid organisms that facilitate the functional analysis of large chromosomal regions and allow the rapid localization of mutations to specific genetic intervals. In mice, well-characterized overlapping deletions are only available at a few chromosomal loci1,2, partly due to drawbacks of existing methods. Here we exploit the random integration of a retrovirus to generate high-resolution sets of nested deletions around defined loci in embryonic stem (ES) cells, with sizes extending from a few kilobases to several megabases. This approach expands the application of Cre-loxP–based chromosome engineering3 because it not only allows the construction of hundreds of overlapping deletions, but also provides molecular entry points to regions based on the retroviral tags. Our approach can be extended to any region of the mouse genome.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Rinchik, E.M. & Russell, L.B. Germ-line deletion mutations in the mouse: tools for intensive functional and physical mapping of regions of the mammalian genome. in Genome Analysis, Vol. 1 (eds Davies, K.E. & Tilghman, S.M.) 121–159(Cold Spring Harbor Laboratory Press, Cold Spring Harbor, 1990).
Cattanach, B.M., Burtenshaw, M.D., Rasberry, C. & Evans, E.P. Large deletions and other gross forms of chromosome imbalance compatible with viability and fertility in the mouse. Nature Genet. 3, 56–61 (1993).
Ramirez-Solis, R., Liu, P. & Bradley, A. Chromosome engineering in mice. Nature 378, 720–724 (1995).
Abuin, A. & Bradley, A. Recycling selectable markers in mouse embryonic stem cells. Mol. Cell. Biol. 16, 1851–1856 (1996).
Zheng, B., Sage, M., Sheppeard, E.A., Jurecic, V. & Bradley, A. Engineering mouse chromosomes with Cre-loxP: range, efficiency and somatic applications. Mol. Cell. Biol. (in press).
Kushi, A., et al. Generation of mutant mice with large chromosomal deletion by use of irradiated ES cells—analysis of large deletion around hprt locus of ES cell. Mamm. Genome 9, 269 –273 (1998).
Thomas, J.W., LaMantia, C. & Magnuson, T. X-ray-induced mutations in mouse embryonic stem cells. Proc. Natl Acad. Sci. USA 95, 1114– 1119 (1998).
Liu, P., Zhang, H., McLellan, A., Vogel, H. & Bradley, A. Embryonic lethality and tumorigenesis caused by segmental aneuploidy on mouse chromosome 11. Genetics 150, 1155–1168 (1998).
You, Y., et al. Chromosomal deletion complexes in mice by radiation of embryonic stem cells. Nature Genet. 15, 285– 288 (1997).
Bridges, C.B. Deficiency. Genetics 2, 445– 465 (1917).
Ashburner, M. Drosophila: A Laboratory Manual(Cold Spring Harbor Press, Cold Spring Harbor, 1989).
Holdener-Kenny, B., Sharan, S.K. & Magnuson, T. Mouse albino-deletions: from genetics to genes in development. Bioessays 14, 831–839 (1992).
Lindsay, E., et al. Congenital heart disease in mice deficient for the DiGeorge syndrome region. Nature 401, 379– 383 (1999).
Friedrich, G. & Soriano, P. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev. 5, 1513–1523 (1991).
Markowitz, D., Goff, S. & Bank, A. A safe packaging line for gene transfer: separating viral genes on two different plasmids. J. Virol. 62, 1120– 1124 (1988).
Soriano, P., Friedrich, G. & Lawinger, P. Promoter interactions in retrovirus vectors introduced into fibroblasts and embryonic stem cells. J. Virol. 65, 2314–2319 (1991).
Hasty, P., Rivera-Perez, J. & Bradley, A. The length of homology required for gene targeting in embryonic stem cells. Mol. Cell. Biol. 11, 5586–5591 (1991)8
Zhang, H., Hasty, P. & Bradley, A. Targeting frequency for deletion vectors in embryonic stem cells. Mol. Cell. Biol. 14, 2404– 2410 (1994).
Ramirez-Solis, R., Davis, A.C. & Bradley, A. Gene targeting in embryonic stem cells. Methods Enzymol. 225, 855–878 (1993).
Dietrich, W., et al. A genetic map of the mouse suitable for typing intraspecific crosses. Genetics 131, 423– 447 (1992).
Ochman, H., Gerber, A.S. & Hartl, D.L. Genetic applications of an inverse polymerase chain reaction. Genetics 120, 621– 623 (1988).
Dalby, B., Pereira, A.J. & Goldstein, L.S. An inverse PCR screen for the detection of P element insertions in cloned genomic intervals in Drosophila melanogaster. Genetics 139, 757–766 ( 1995).
Cai, W.W., Reneker, J., Chow, C.W., Vaishnav, M. & Bradley, A. An anchored framework BAC map of mouse chromosome 11 assembled using multiplex oligonucleotide hybridization. Genomics 54, 387–397 ( 1998).
Dietrich, W.F. et al. A comprehensive genetic map of the mouse genome. Nature 380, 149–152 ( 1996).
Acknowledgements
We thank N. Copeland and N. Jenkins for CAST/Ei mice; P. Liu for Hsd17b1 targeting vectors; W. Cai and J. Li for advice on BAC screening; Y. Cheah for generating chimaeras; S. Perez for preparation of the manuscript; and G. Luo, B. Zheng, P. Biggs, D. Thompson and M. Wentland for critical reading of the manuscript. This work was supported by grants from The National Cancer Institute. X.W. is an Associate and A.B. an Investigator with the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Su, H., Wang, X. & Bradley, A. Nested chromosomal deletions induced with retroviral vectors in mice . Nat Genet 24, 92–95 (2000). https://doi.org/10.1038/71756
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/71756
This article is cited by
-
A transposon-based chromosomal engineering method to survey a large cis-regulatory landscape in mice
Nature Genetics (2009)
-
A retroviral strategy that efficiently creates chromosomal deletions in mammalian cells
Nature Methods (2007)
-
The art and design of genetic screens: mouse
Nature Reviews Genetics (2005)
-
X-ray-induced deletion complexes in embryonic stem cells on mouse chromosome 15
Mammalian Genome (2005)
-
Allelic phasing of a mouse chromosome 11 deficiency influences p53 tumorigenicity
Oncogene (2003)