Abstract
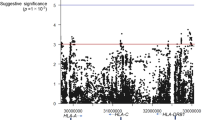
Hypovolemic shock (dengue shock syndrome (DSS)) is the most common life-threatening complication of dengue. We conducted a genome-wide association study of 2,008 pediatric cases treated for DSS and 2,018 controls from Vietnam. Replication of the most significantly associated markers was carried out in an independent Vietnamese sample of 1,737 cases and 2,934 controls. SNPs at two loci showed genome-wide significant association with DSS. We identified a susceptibility locus at MICB (major histocompatibility complex (MHC) class I polypeptide-related sequence B), which was within the broad MHC region on chromosome 6 but outside the class I and class II HLA loci (rs3132468, Pmeta = 4.41 × 10−11, per-allele odds ratio (OR) = 1.34 (95% confidence interval: 1.23–1.46)). We identified associated variants within PLCE1 (phospholipase C, epsilon 1) on chromosome 10 (rs3765524, Pmeta = 3.08 × 10−10, per-allele OR = 0.80 (95% confidence interval: 0.75–0.86)). We identify two loci associated with susceptibility to DSS in people with dengue, suggesting possible mechanisms for this severe complication of dengue.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
Accession codes
References
Gubler, D.J. Dengue and dengue hemorrhagic fever. Clin. Microbiol. Rev. 11, 480–496 (1998).
Anders, K.L. et al. Epidemiological factors associated with dengue shock syndrome and mortality in hospitalized dengue patients in Ho Chi Minh City, Vietnam. Am. J. Trop. Med. Hyg. 84, 127–134 (2011).
Thai, K.T. et al. Seroprevalence of dengue antibodies, annual incidence and risk factors among children in southern Vietnam. Trop. Med. Int. Health 10, 379–386 (2005).
Loke, H. et al. Susceptibility to dengue hemorrhagic fever in vietnam: evidence of an association with variation in the vitamin D receptor and FC gamma receptor IIA genes. Am. J. Trop. Med. Hyg. 67, 102–106 (2002).
Loke, H. et al. Strong HLA class I–restricted T cell responses in dengue hemorrhagic fever: a double-edged sword? J. Infect. Dis. 184, 1369–1373 (2001).
Stephens, H.A. et al. HLA-A and -B allele associations with secondary dengue virus infections correlate with disease severity and the infecting viral serotype in ethnic Thais. Tissue Antigens 60, 309–318 (2002).
Sakuntabhai, A. et al. A variant in the CD209 promoter is associated with severity of dengue disease. Nat. Genet. 37, 507–513 (2005).
Vejbaesya, S. et al. TNF and LTA gene, allele and extended HLA haplotype associations with severe dengue virus infection in ethnic Thais. J. Infect. Dis. 199, 1442–1448 (2009).
Steinle, A. et al. Interactions of human NKG2D with its ligands MICA, MICB, and homologs of the mouse RAE-1 protein family. Immunogenetics 53, 279–287 (2001).
González, S., Lopez-Soto, A., Suarez-Alvarez, B., Lopez-Vazquez, A. & Lopez-Larrea, C. NKG2D ligands: key targets of the immune response. Trends Immunol. 29, 397–403 (2008).
Garrity, D., Call, M.E., Feng, J. & Wucherpfennig, K.W. The activating NKG2D receptor assembles in the membrane with two signaling dimers into a hexameric structure. Proc. Natl. Acad. Sci. USA 102, 7641–7646 (2005).
Hoang, L.T. et al. The early whole-blood transcriptional signature of dengue virus and features associated with progression to dengue shock syndrome in Vietnamese children and young adults. J. Virol. 84, 12982–12994 (2010).
Libraty, D.H. et al. Differing influences of virus burden and immune activation on disease severity in secondary dengue-3 virus infections. J. Infect. Dis. 185, 1213–1221 (2002).
Vaughn, D.W. et al. Dengue viremia titer, antibody response pattern, and virus serotype correlate with disease severity. J. Infect. Dis. 181, 2–9 (2000).
Kumar, V. et al. Genome-wide association study identifies a susceptibility locus for HCV-induced hepatocellular carcinoma. Nat. Genet. 43, 455–458 (2011).
Hinkes, B. et al. Positional cloning uncovers mutations in PLCE1 responsible for a nephrotic syndrome variant that may be reversible. Nat. Genet. 38, 1397–1405 (2006).
Chau, T.N. et al. Dengue virus infections and maternal antibody decay in a prospective birth cohort study of Vietnamese infants. J. Infect. Dis. 200, 1893–1900 (2009).
Tien, N.T. et al. A prospective cohort study of dengue infection in schoolchildren in Long Xuyen, Viet Nam. Trans. R. Soc. Trop. Med. Hyg. 104, 592–600 (2010).
Balmaseda, A. et al. Trends in patterns of dengue transmission over 4 years in a pediatric cohort study in Nicaragua. J. Infect. Dis. 201, 5–14 (2010).
Porter, K.R. et al. Epidemiology of dengue and dengue hemorrhagic fever in a cohort of adults living in Bandung, West Java, Indonesia. Am. J. Trop. Med. Hyg. 72, 60–66 (2005).
Endy, T.P. et al. Epidemiology of inapparent and symptomatic acute dengue virus infection: a prospective study of primary school children in Kamphaeng Phet, Thailand. Am. J. Epidemiol. 156, 40–51 (2002).
Mammen, M.P. et al. Spatial and temporal clustering of dengue virus transmission in Thai villages. PLoS Med. 5, e205 (2008).
Endy, T.P. et al. Determinants of inapparent and symptomatic dengue infection in a prospective study of primary school children in Kamphaeng Phet, Thailand. PLoS Negl. Trop. Dis. 5, e975 (2011).
Mells, G.F. et al. Genome-wide association study identifies 12 new susceptibility loci for primary biliary cirrhosis. Nat. Genet. 43, 329–332 (2011).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Price, A.L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006).
Purdue, M.P. et al. Genome-wide association study of renal cell carcinoma identifies two susceptibility loci on 2p21 and 11q13.3. Nat. Genet. 43, 60–65 (2011).
Thomas, G. et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1). Nat. Genet. 41, 579–584 (2009).
Lettre, G., Lange, C. & Hirschhorn, J.N. Genetic model testing and statistical power in population-based association studies of quantitative traits. Genet. Epidemiol. 31, 358–362 (2007).
McGovern, D.P. et al. Genome-wide association identifies multiple ulcerative colitis susceptibility loci. Nat. Genet. 42, 332–337 (2010).
Asano, K. et al. A genome-wide association study identifies three new susceptibility loci for ulcerative colitis in the Japanese population. Nat. Genet. 41, 1325–1329 (2009).
Barrett, J.C., Fry, B., Maller, J. & Daly, M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–265 (2005).
Acknowledgements
This work was supported by the Wellcome Trust, United Kingdom (grant 088791/A/09/Z and 084368/Z/07/Z), and the Agency for Science, Technology and Research, Singapore.
Author information
Authors and Affiliations
Contributions
M.L.H. and C.P.S. are the study principal investigators who conceived and obtained funding for the project. C.C.K. organized and supervised the GWAS and replication genotyping pipeline, devised the overall analysis plan and wrote the first draft of the manuscript with input from M.L.H., C.P.S. and S.D. T.N.B.C. is the lead coordinator of clinical samples and phenotypes for both the discovery and replication stages. J.P. and D.E.K.T. performed genotyping and quality checks on all samples. C.C.K., S.D., R.T.H.O. and Y.-Y.T. analyzed the data. H.T.L., S.J.D., B.W., J.F., T.V.T., T.T.G., N.T.N.B., L.T.T., L.B.L., N.M.T., N.T.H.T., M.N.L., N.M.N., N.T.H., N.V.V.C., T.T.T. and A.S. coordinated and contributed patient and database phenotype collections as lead investigators for their respective sample collections. D.E.K.T. and J.P. performed genotyping and DNA quality checks. All authors critically reviewed manuscript revisions and contributed intellectual input to the final submission.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5 and Supplementary Tables 1–3 (PDF 2865 kb)
Rights and permissions
About this article
Cite this article
Khor, C., Chau, T., Pang, J. et al. Genome-wide association study identifies susceptibility loci for dengue shock syndrome at MICB and PLCE1. Nat Genet 43, 1139–1141 (2011). https://doi.org/10.1038/ng.960
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.960
This article is cited by
-
Complement receptor type 1 and 2 (CR1 and CR2) gene polymorphisms and plasma protein levels are associated with the Dengue disease severity
Scientific Reports (2023)
-
Differential expression of microRNA, miR-150 and enhancer of zeste homolog 2 (EZH2) in peripheral blood cells as early prognostic markers of severe forms of dengue
Journal of Biomedical Science (2020)
-
Complement protein levels and MBL2 polymorphisms are associated with dengue and disease severity
Scientific Reports (2020)
-
Replicated methylation changes associated with eczema herpeticum and allergic response
Clinical Epigenetics (2019)
-
Metabolic perturbations and cellular stress underpin susceptibility to symptomatic live-attenuated yellow fever infection
Nature Medicine (2019)