Abstract
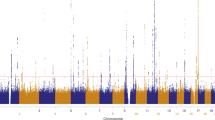
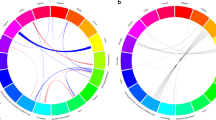
We performed a genome-wide association study of melanoma in a discovery cohort of 2,168 Australian individuals with melanoma and 4,387 control individuals. In this discovery phase, we confirm several previously characterized melanoma-associated loci at MC1R, ASIP and MTAP–CDKN2A. We selected variants at nine loci for replication in three independent case-control studies (Europe: 2,804 subjects with melanoma, 7,618 control subjects; United States 1: 1,804 subjects with melanoma, 1,026 control subjects; United States 2: 585 subjects with melanoma, 6,500 control subjects). The combined meta-analysis of all case-control studies identified a new susceptibility locus at 1q21.3 (rs7412746, P = 9.0 × 10−11, OR in combined replication cohorts of 0.89 (95% CI 0.85–0.95)). We also show evidence suggesting that melanoma associates with 1q42.12 (rs3219090, P = 9.3 × 10−8). The associated variants at the 1q21.3 locus span a region with ten genes, and plausible candidate genes for melanoma susceptibility include ARNT and SETDB1. Variants at the 1q21.3 locus do not seem to be associated with human pigmentation or measures of nevus density.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Bishop, D.T. et al. Genome-wide association study identifies three loci associated with melanoma risk. Nat. Genet. 41, 920–925 (2009).
Brown, K.M. et al. Common sequence variants on 20q11.22 confer melanoma susceptibility. Nat. Genet. 40, 838–840 (2008).
Gudbjartsson, D.F. et al. ASIP and TYR pigmentation variants associate with cutaneous melanoma and basal cell carcinoma. Nat. Genet. 40, 886–891 (2008).
Falchi, M. et al. Genome-wide association study identifies variants at 9p21 and 22q13 associated with development of cutaneous nevi. Nat. Genet. 41, 915–919 (2009).
Duffy, D.L. et al. IRF4 variants have age-specific effects on nevus count and predispose to melanoma. Am. J. Hum. Genet. 87, 6–16 (2010).
Baxter, A.J. et al. The Queensland Study of Melanoma: environmental and genetic associations (Q-MEGA); study design, baseline characteristics, and repeatability of phenotype and sun exposure measures. Twin Res. Hum. Genet. 11, 183–196 (2008).
Cust, A.E. et al. Population-based, case-control-family design to investigate genetic and environmental influences on melanoma risk: Australian Melanoma Family Study. Am. J. Epidemiol. 170, 1541–1554 (2009).
Painter, J.N. et al. Genome-wide association study identifies a locus at 7p15.2 associated with endometriosis. Nat. Genet. 43, 51–54 (2010).
Durbin, R.M. et al. A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073 (2010).
Li, C. et al. Genetic variants of the ADPRT, XRCC1 and APE1 genes and risk of cutaneous melanoma. Carcinogenesis 27, 1894–1901 (2006).
Johansson, P., Pavey, S. & Hayward, N. Confirmation of a BRAF mutation-associated gene expression signature in melanoma. Pigment Cell Res. 20, 216–221 (2007).
Ceol, C.J. et al. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature 471, 513–517 (2011).
Zhang, M., Qureshi, A.A., Guo, Q. & Han, J. Genetic variation in DNA repair pathway genes and melanoma risk. DNA Repair (Amst). 10, 111–116 (2011).
Park, J.H. et al. Estimation of effect size distribution from genome-wide association studies and implications for future discoveries. Nat. Genet. 42, 570–575 (2010).
Price, A.L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006).
Li, Y. & Mach Abecasis, G. 1.0: Rapid Haplotype Reconstruction and Missing Genotype Inference. Am. J. Hum. Genet. S79, 2290 (2006).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Queensland Cancer Registry. Cancer in Queensland: incidence and mortality, 1982 to 2005 (The Cancer Council Queensland, 2008).
Do, K.A., Aitken, J.F., Green, A.C. & Martin, N.G. Analysis of melanoma onset: assessing familial aggregation by using estimating equations and fitting variance components via Bayesian random effects models. Twin Res. 7, 98–113 (2004).
James, J.W. Frequency in relatives for an all-or-none trait. Ann. Hum. Genet. 35, 47–49 (1971).
Pruim, R.J. et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 26, 2336–2337 (2010).
Acknowledgements
The AMFS and Q-MEGA gratefully acknowledge all their participants and the hard work of all their research interviewers and examiners and especially acknowledge C. Agha-Hamilton for managing the AMFS biospecimens. Q-MEGA thanks A. Baxter, M. de Nooyer, I. Gardner, D. Statham, B. Haddon, M.J. Wright, J. Palmer, J. Symmons, B. Castellano, L. Bardsley, S. Smith, D. Smyth, L. Wallace, M.J. Campbell, A. Caracella, M. Kvaskoff, O. Zheng, B. Chapman and H. Beeby for their input in project management, sample processing and database development. We are grateful to the many research assistants and interviewers for assistance with the studies contributing to the QIMR and AMFS collections. We acknowledge with appreciation all the participants in the QIMR and endometriosis studies. We thank Endometriosis Associations for supporting study recruitment. We thank S. Nicolaides and the Queensland Medical Laboratory for the pro bono collection and delivery of blood samples and other pathology services for assistance with blood collection. We thank the following US state cancer registries for their help: Alabama, Arizona, Arkansas, California, Colorado, Connecticut, Delaware, Florida, Georgia, Idaho, Illinois, Indiana, Iowa, Kentucky, Louisiana, Maine, Maryland, Massachusetts, Michigan, Nebraska, New Hampshire, New Jersey, New York, North Carolina, North Dakota, Ohio, Oklahoma, Oregon, Pennsylvania, Rhode Island, South Carolina, Tennessee, Texas, Virginia, Washington and Wyoming.
This work was supported by the Melanoma Research Alliance, the National Cancer Institute (NCI) of the US National Institutes of Health (NIH) (CA88363, CA83115, CA122838, CA87969, CA055075, CA100264, CA133996 and CA49449), the National Health and Medical Research Council (NHMRC) of Australia (107359, 200071, 241944, 339462, 380385, 389927, 389875, 389891, 389892, 389938, 402761, 443036, 442915, 442981, 496610, 496675, 496739, 552485 and 552498), the Cancer Councils New South Wales, Victoria and Queensland, the Cancer Institute New South Wales, the Cooperative Research Centre (CRC) for Discovery of Genes for Common Human Diseases, Cerylid Biosciences (Melbourne), the Australian Cancer Research Foundation, The Wellcome Trust and donations from Neville and Shirley Hawkins. Endometriosis sample genotyping was funded by a grant from the Wellcome Trust (WT084766/Z/08/Z). The Australian Twin Registry is supported by an Australian National Health and Medical Research Council (NHMRC) Enabling Grant (2004–2009). D.L.D., N.K.H. and G.W.M. are supported by the NHMRC Fellowships scheme and J.L.H. is an Australia Fellow of the NHMRC. S.M. is the recipient of a Career Development Award from the NHMRC (496674, 613705). D.C.W. is a Future Fellow of the Australian Research Council. K.M.B. is supported by the NCI of the NIH. J.M.T. is supported by the NCI of the NIH (CA109544). B.K.A. is supported by the University of Sydney Medical Foundation. A.E.C. is the recipient of a NHMRC public health postdoctoral fellowship (520018) and a Cancer Institute NSW Early Career Development Fellowship (10/ECF/2-06). A.M.G. and M.T.L. were supported by the Intramural Research Program of the NIH, NCI and DCEG. The GenoMEL replication sample was funded by the European Commission under the 6th Framework Programme (contract no. LSHC-CT-2006-018702), by Cancer Research UK Programme Award (C588/A4994) and by the NIH (CA83115). Research at M.D. Anderson Cancer Center (MDACC) was partially supported by NIH grants (CA100264, 2P50CA093459, P30CA016672 and R01CA133996). The Center for Inherited Disease Research performed genotyping for MDACC and is supported by contract (HHSN268200782096C). GENEVA performed data cleaning and is supported by an NIH grant (HG004446). Research by the Barcelona team was supported by Fondo de Investigaciones Sanitarias (09/1393).
Author information
Authors and Affiliations
Contributions
S.M., N.K.H. and K.M.B. wrote the manuscript. G.W.M., N.K.H., K.M.B., J.M.T., A.K.H., Z.Z.Z. and M.S. designed, analyzed and managed the sample preparation and genotyping aspects of the study. S.M. and J.Z.L. performed data analysis. D.C.W., D.L.D., G.W.M., N.K.H., S.M., J.N.P., D.R.N. and N.G.M. oversaw collection of the Queensland samples and contributed to statistical analyses, data interpretation and manuscript preparation. G.J.M., A.E.C., E.A.H., H.S., J.A.M., J.J., M.F., M.A.J., R.F.K., G.G.G., B.K.A., J.F.A. and J.L.H. oversaw sample collection, genotyping and analysis in the AMFS study. D.T.B., J.A.N.-B., M.M.I., H.O., S.P., G.B.-S., J.H., F.D., M.T.L., T.D., R.M., E.A., B.B.-d.P., A.M.G., P.A.K., N.A.G., P.H. and D.E.E. contributed to the collection, genotyping and analysis of the GenoMEL samples. C.I.A., Q.W., L.-E.W. and J.E.L. contributed to the collection, genotyping and analysis of the United States 1–M.D. Anderson Cancer Center samples. A.A.Q., M.Z. and J.H. contributed to the collection, genotyping and analysis of the United States 2–Harvard samples.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1 and 2, Supplementary Figures 1–5 and Supplementary Note (PDF 530 kb)
Rights and permissions
About this article
Cite this article
MacGregor, S., Montgomery, G., Liu, J. et al. Genome-wide association study identifies a new melanoma susceptibility locus at 1q21.3. Nat Genet 43, 1114–1118 (2011). https://doi.org/10.1038/ng.958
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.958
This article is cited by
-
Implementation of individualised polygenic risk score analysis: a test case of a family of four
BMC Medical Genomics (2022)
-
Massively parallel reporter assays of melanoma risk variants identify MX2 as a gene promoting melanoma
Nature Communications (2020)
-
Genome-wide association meta-analyses combining multiple risk phenotypes provide insights into the genetic architecture of cutaneous melanoma susceptibility
Nature Genetics (2020)
-
PARP1 rs1805407 Increases Sensitivity to PARP1 Inhibitors in Cancer Cells Suggesting an Improved Therapeutic Strategy
Scientific Reports (2019)
-
AKT methylation by SETDB1 promotes AKT kinase activity and oncogenic functions
Nature Cell Biology (2019)