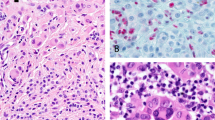
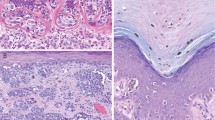
Abstract
Common acquired melanocytic nevi are benign neoplasms that are composed of small, uniform melanocytes and are typically present as flat or slightly elevated pigmented lesions on the skin. We describe two families with a new autosomal dominant syndrome characterized by multiple, skin-colored, elevated melanocytic tumors. In contrast to common acquired nevi, the melanocytic neoplasms in affected family members ranged histopathologically from epithelioid nevi to atypical melanocytic proliferations that showed overlapping features with melanoma. Some affected individuals developed uveal or cutaneous melanomas. Segregating with this phenotype, we found inactivating germline mutations of BAP1, which encodes a ubiquitin carboxy-terminal hydrolase. The majority of melanocytic neoplasms lost the remaining wild-type allele of BAP1 by various somatic alterations. In addition, we found BAP1 mutations in a subset of sporadic melanocytic neoplasms showing histological similarities to the familial tumors. These findings suggest that loss of BAP1 is associated with a clinically and morphologically distinct type of melanocytic neoplasm.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Palmedo, G. et al. The T1796A mutation of the BRAF gene is absent in Spitz nevi. J. Cutan. Pathol. 31, 266–270 (2004).
Knudson, A.G. Jr. Mutation and cancer: statistical study of retinoblastoma. Proc. Natl. Acad. Sci. USA 68, 820–823 (1971).
Gnirke, A. et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat. Biotechnol. 27, 182–189 (2009).
Harbour, J.W. et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science 330, 1410–1413 (2010).
Van Raamsdonk, C.D. et al. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature 457, 599–602 (2009).
Van Raamsdonk, C.D. et al. Mutations in GNA11 in uveal melanoma. N. Engl. J. Med. 363, 2191–2199 (2010).
Matsuoka, S. et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316, 1160–1166 (2007).
Stokes, M.P. et al. Profiling of UV-induced ATM/ATR signaling pathways. Proc. Natl. Acad. Sci. USA 104, 19855–19860 (2007).
Ventii, K.H. et al. BRCA1-associated protein-1 is a tumor suppressor that requires deubiquitinating activity and nuclear localization. Cancer Res. 68, 6953–6962 (2008).
Scheuermann, J.C. et al. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature 465, 243–247 (2010).
Jensen, D.E. et al. BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene 16, 1097–1112 (1998).
Wood, L.D. et al. The genomic landscapes of human breast and colorectal cancers. Science 318, 1108–1113 (2007).
Buchhagen, D.L., Qiu, L. & Etkind, P. Homozygous deletion, rearrangement and hypermethylation implicate chromosome region 3p14.3–3p21.3 in sporadic breast-cancer development. Int. J. Cancer 57, 473–479 (1994).
Curtin, J.A. et al. Distinct sets of genetic alterations in melanoma. N. Engl. J. Med. 353, 2135–2147 (2005).
Ng, P.C. & Henikoff, S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 31, 3812–3814 (2003).
Ramensky, V., Bork, P. & Sunyaev, S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 30, 3894–3900 (2002).
Reva, B., Antipin, Y. & Sander, C. Determinants of protein function revealed by combinatorial entropy optimization. Genome Biol. 8, R232 (2007).
Acknowledgements
We are indebted to all participating subjects for their enthusiastic participation in the study. We thank W. Stieber for clinical photography, U. Schmidbauer for providing histotechnical services, M. Leversha for performing FISH, A. Heguy for supporting sequencing, M. Asher for immunohistochemistry, H. Al-Ahmadie for assistance with the histological images and P. Dillinger for helping to collect the tissue samples. T.W. was supported by a Max-Kade Fellowship and expresses special thanks to H. Kerl for supporting, encouraging and inspiring his academic interests. A.C.O. is supported by the Austrian Science Fund (J3013) and K.G.G. by the Deutsche Forschungsgemeinschaft (GR 3671/1-1a). This work was funded by grants from the US National Institutes of Health (R01 CA131524), Geoffrey Beene Cancer Research Center (CC 66270), the American Skin Association (all to B.C.B.), the Andrew Sabin Family Foundation (to K.O.), the European Commission (GENINCA, contract no. HEALTH-F2-2008-202230) and the Jubilaeumsfonds of the Oesterreichische Nationalbank (13837).
Author information
Authors and Affiliations
Contributions
Project planning and experimental design: T.W., B.C.B. and M.R.S. Review of clinical phenotypes: T.W., I.F., I.W. and J.C.B. Review of histology and immunohistology: T.W., B.C.B., H.K., R.M., L.C., I.F., A.R. and A.O. FISH analysis: T.W. and G.P. Sample collection: T.W., H.K., W.W., K.O. and D.A. aCGH: T.W. and A.C.O. Linkage analysis: A.C.O. and C.W. Mutation analysis: T.W., P.U. and S.L. Next-generation sequencing and data analysis: T.W., A.V., A.E.L., N.D.S. and M.P. Manuscript writing: T.W., B.C.B., R.M., M.R.S. and K.G.G. Revision of the manuscript: all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–15 and Supplementary Tables 1–9 (PDF 11936 kb)
Rights and permissions
About this article
Cite this article
Wiesner, T., Obenauf, A., Murali, R. et al. Germline mutations in BAP1 predispose to melanocytic tumors. Nat Genet 43, 1018–1021 (2011). https://doi.org/10.1038/ng.910
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.910
This article is cited by
-
The BAP1 nuclear deubiquitinase is involved in the nonhomologous end-joining pathway of double-strand DNA repair through interaction with DNA-PK
Oncogene (2024)
-
Deficiency of BAP1 inhibits neuroblastoma tumorigenesis through destabilization of MYCN
Cell Death & Disease (2023)
-
Two unique BAP1 pathogenic variants identified in the same family by panel cascade testing
Familial Cancer (2023)
-
ITGB2-ICAM1 axis promotes liver metastasis in BAP1-mutated uveal melanoma with retained hypoxia and ECM signatures
Cellular Oncology (2023)
-
Analysis of uveal melanomas and paired constitutional DNA for exclusion of a BAP1-tumor predisposition syndrome
Familial Cancer (2023)