Abstract
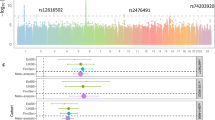
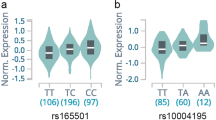
Graves' disease is a common autoimmune disorder characterized by thyroid stimulating hormone receptor autoantibodies (TRAb) and hyperthyroidism. To investigate the genetic architecture of Graves' disease, we conducted a genome-wide association study in 1,536 individuals with Graves' disease (cases) and 1,516 controls. We further evaluated a group of associated SNPs in a second set of 3,994 cases and 3,510 controls. We confirmed four previously reported loci (in the major histocompatibility complex, TSHR, CTLA4 and FCRL3) and identified two new susceptibility loci (the RNASET2-FGFR1OP-CCR6 region at 6q27 (Pcombined = 6.85 × 10−10 for rs9355610) and an intergenic region at 4p14 (Pcombined = 1.08 × 10−13 for rs6832151)). These newly associated SNPs were correlated with the expression levels of RNASET2 at 6q27, of CHRNA9 and of a previously uncharacterized gene at 4p14, respectively. Moreover, we identified strong associations of TSHR and major histocompatibility complex class II variants with persistently TRAb-positive Graves' disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Simmonds, M.J. et al. Regression mapping of association between the human leukocyte antigen region and Graves disease. Am. J. Hum. Genet. 76, 157–163 (2005).
Simmonds, M.J. et al. A novel and major association of HLA-C in Graves' disease that eclipses the classical HLA-DRB1 effect. Hum. Mol. Genet. 16, 2149–2153 (2007).
Ueda, H. et al. Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature 423, 506–511 (2003).
Zhao, S.X. et al. Association of the CTLA4 gene with Graves' disease in the Chinese Han population. PLoS ONE 5, e9821 (2010).
Brand, O.J. et al. Association of the thyroid stimulating hormone receptor gene (TSHR) with Graves' disease. Hum. Mol. Genet. 18, 1704–1713 (2009).
Hiratani, H. et al. Multiple SNPs in intron 7 of thyrotropin receptor are associated with Graves' disease. J. Clin. Endocrinol. Metab. 90, 2898–2903 (2005).
Velaga, M.R. et al. The codon 620 tryptophan allele of the lymphoid tyrosine phosphatase (LYP) gene is a major determinant of Graves' disease. J. Clin. Endocrinol. Metab. 89, 5862–5865 (2004).
Burton, P.R. et al. Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat. Genet. 39, 1329–1337 (2007).
Chistiakov, D.A. & Chistiakov, A.P. Is FCRL3 a new general autoimmunity gene? Hum. Immunol. 68, 375–383 (2007).
Kochi, Y. et al. A functional variant in FCRL3, encoding Fc receptor-like 3, is associated with rheumatoid arthritis and several autoimmunities. Nat. Genet. 37, 478–485 (2005).
Kamatani, Y. et al. A genome-wide association study identifies variants in the HLA-DP locus associated with chronic hepatitis B in Asians. Nat. Genet. 41, 591–595 (2009).
Dixon, A.L. et al. A genome-wide association study of global gene expression. Nat. Genet. 39, 1202–1207 (2007).
Zeller, T. et al. Genetics and beyond – the transcriptome of human monocytes and disease susceptibility. PLoS ONE 5, e10693 (2010).
Barrett, J.C. et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat. Genet. 40, 955–962 (2008).
Kochi, Y. et al. A regulatory variant in CCR6 is associated with rheumatoid arthritis susceptibility. Nat. Genet. 42, 515–519 (2010).
Stahl, E.A. et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat. Genet. 42, 508–514 (2010).
Jin, Y. et al. Common variants in FOXP1 are associated with generalized vitiligo. Nat. Genet. 42, 576–578 (2010).
Quan, C. et al. Genome-wide association study for vitiligo identifies susceptibility loci at 6q27 and the MHC. Nat. Genet. 42, 614–618 (2010).
Acquati, F. et al. Tumor and metastasis suppression by the human RNASET2 gene. Int. J. Oncol. 26, 1159–1168 (2005).
Liu, J. et al. Chromosome 6 encoded RNaseT2 protein is a cell growth regulator. J. Cell Mol. Med. 14, 1146–1155 (2010).
Everts, B. et al. Omega-1, a glycoprotein secreted by Schistosoma mansoni eggs, drives Th2 responses. J. Exp. Med. 206, 1673–1680 (2009).
Steinfelder, S. et al. The major component in schistosome eggs responsible for conditioning dendritic cells for Th2 polarization is a T2 ribonuclease (omega-1). J. Exp. Med. 206, 1681–1690 (2009).
Feldt-Rasmussen, U., Schleusener, H. & Carayon, P. Meta-analysis evaluation of the impact of thyrotropin receptor antibodies on long term remission after medical therapy of Graves' disease. J. Clin. Endocrinol. Metab. 78, 98–102 (1994).
Cappelli, C. et al. Prognostic value of thyrotropin receptor antibodies (TRAb) in Graves' disease: a 120 months prospective study. Endocr. J. 54, 713–720 (2007).
Song, H.D. et al. Functional SNPs in the SCGB3A2 promoter are associated with susceptibility to Graves' disease. Hum. Mol. Genet. 18, 1156–1170 (2009).
Price, A.L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Breslow, N.E. & Day, N.E. Statistical methods in cancer research. Volume I - the analysis of case-control studies. IARC Sci. Publ. 32, 5–338 (1980).
Li, Y., Willer, C., Sanna, S. & Abecasis, G. Genotype imputation. Annu. Rev. Genomics Hum. Genet. 10, 387–406 (2009).
Frazer, K.A. et al. A second generation human haplotype map of over 3.1 million SNPs. Nature 449, 851–861 (2007).
Aulchenko, Y.S., Struchalin, M. & van Duijn, C. ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinformatics 11, 134 (2010).
Carella, C. et al. Serum thyrotropin receptor antibodies concentrations in patients with Graves' disease before, at the end of methimazole treatment, and after drug withdrawal: evidence that the activity of thyrotropin receptor antibody and/or thyroid response modify during the observation period. Thyroid 16, 295–302 (2006).
Acknowledgements
We thank all subjects for participating in this study. This work was supported in part by the National Natural Science Foundation of China (30971595, 30971383 and 31000556), Chinese National Natural Science Fund for Distinguished Young Scholars (30625019), Chinese High-Tech Program (2007AA02Z175 and 2009AA022709), Shanghai Science and Technology Committee (10JC1410400, 09DZ2291900, 09JC1411300 and 10ZR1421400) and Program for Innovative Research Team of Shanghai Municipal Education Committee.
Author information
Authors and Affiliations
Consortia
Contributions
Z.C. was responsible for the coordination of the project. The writing team consisted of H.-D.S., X.C., W.H., S.-X.Z., J.-L.C., S.-J.C. and Z.C. Z.C., H.-D.S., W.H., G.N., J.-L.C. and S.-J.C. contributed to the study design. H.-D.S., W.H., C.-M.P. and X.C. contributed to the project management. H.-D.S., J.L., G.-Q.G., X.-M.Z., C.-M.P., G.-Y.Y., C.-G.L., L.-Q.X., W.L., S.-Y.Y., S.-X.Z., W.-H.D., C.-L.Z., B.-L.L., X.-N.Z., F.S., Z.-Q.W., Z.-Y.S., C.-Y.Z., W.-H.S., H.-M.C., J.-H.M., B.H., P.L., H.J., C.-C.G., M.Z., L.-B.L., G.C., Q.S., Y.-D.P. and J.-J.Z. took part in the collection of clinical samples and DNA and sample quality control. W.H., H.-F.W., M.S., X.C., F.X., W.-W.S. and J.-X.S. contributed to whole-genome scan genotyping. C.-M.P., J.-Y.S., L.-Q.X., W.L., S.-Y.Y., S.-X.Z., T.G., X.C., M.S., F.X. and W.-W.S. took part in replication genotyping. S.-X.Z., C.-M.P., M.Z., B.-L.L., C.-C.G., Z.-H.G., W.L., S.-Y.Y., L.-Q.X. and Q.-H.H. contributed to cloning of GDCG4p14 and real-time RT-PCR. X.C., W.H., J.-X.S., H.-D.S., S.-X.Z., Z.-H.G., J.-L.C., S.-J.C. and Z.C. took part in the statistical analysis. S.-X.Z. and L.L. performed imputation and cis-eQTL analysis.
Corresponding authors
Ethics declarations
Competing interests
The author declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Note, Supplementary Figures 1–9 and Supplementary Tables 1–11. (PDF 1820 kb)
Rights and permissions
About this article
Cite this article
The China Consortium for the Genetics of Autoimmune Thyroid Disease. A genome-wide association study identifies two new risk loci for Graves' disease. Nat Genet 43, 897–901 (2011). https://doi.org/10.1038/ng.898
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.898
This article is cited by
-
Shared genetic architecture between autoimmune disorders and B-cell acute lymphoblastic leukemia: insights from large-scale genome-wide cross-trait analysis
BMC Medicine (2024)
-
Deficiency of the HGF/Met pathway leads to thyroid dysgenesis by impeding late thyroid expansion
Nature Communications (2024)
-
The genetics of Graves’ disease
Reviews in Endocrine and Metabolic Disorders (2024)
-
Cross-disorder genetic analysis of immune diseases reveals distinct gene associations that converge on common pathways
Nature Communications (2023)
-
Markers of immune dysregulation in response to the ageing gut: insights from aged murine gut microbiota transplants
BMC Gastroenterology (2022)