Abstract
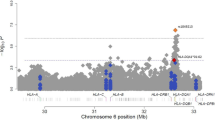
Asthma is a common disease with a complex risk architecture including both genetic and environmental factors. We performed a meta-analysis of North American genome-wide association studies of asthma in 5,416 individuals with asthma (cases) including individuals of European American, African American or African Caribbean, and Latino ancestry, with replication in an additional 12,649 individuals from the same ethnic groups. We identified five susceptibility loci. Four were at previously reported loci on 17q21, near IL1RL1, TSLP and IL33, but we report for the first time, to our knowledge, that these loci are associated with asthma risk in three ethnic groups. In addition, we identified a new asthma susceptibility locus at PYHIN1, with the association being specific to individuals of African descent (P = 3.9 × 10−9). These results suggest that some asthma susceptibility loci are robust to differences in ancestry when sufficiently large samples sizes are investigated, and that ancestry-specific associations also contribute to the complex genetic architecture of asthma.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Masoli, M., Fabian, D., Holt, S. & Beasley, R. The global burden of asthma: executive summary of the GINA Dissemination Committee report. Allergy 59, 469–478 (2004).
Moorman, J.E. et al. National surveillance for asthma–United States, 1980–2004. MMWR Surveill. Summ. 56, 1–54 (2007).
Duffy, D.L., Martin, N.G., Battistutta, D., Hopper, J.L. & Mathews, J.D. Genetics of asthma and hay fever in Australian twins. Am. Rev. Respir. Dis. 142, 1351–1358 (1990).
Nieminen, M.M., Kaprio, J. & Koskenvuo, M. A population-based study of bronchial asthma in adult twin pairs. Chest 100, 70–75 (1991).
Ober, C. & Hoffjan, S. Asthma genetics 2006: the long and winding road to gene discovery. Genes Immun. 7, 95–100 (2006).
Vercelli, D. Discovering susceptibility genes for asthma and allergy. Nat. Rev. Immunol. 8, 169–182 (2008).
Moffatt, M.F. et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature 448, 470–473 (2007).
Himes, B.E. et al. Genome-wide association analysis identifies PDE4D as an asthma-susceptibility gene. Am. J. Hum. Genet. 84, 581–593 (2009).
Li, X. et al. Genome-wide association study of asthma identifies RAD50-IL13 and HLA-DR/DQ regions. J. Allergy Clin. Immunol. 125, 328–335 e11 (2010).
Sleiman, P.M. et al. Variants of DENND1B associated with asthma in children. N. Engl. J. Med. 362, 36–44 (2010).
Hancock, D.B. et al. Genome-wide association study implicates chromosome 9q21.31 as a susceptibility locus for asthma in Mexican children. PLoS Genet. 5, e1000623 (2009).
Choudhry, S. et al. Genome-wide screen for asthma in Puerto Ricans: evidence for association with 5q23 region. Hum. Genet. 123, 455–468 (2008).
Mathias, R.A. et al. A genome-wide association study on African-ancestry populations for asthma. J. Allergy Clin. Immunol. 125, 336–346 e4 (2010).
Bouzigon, E. et al. Effect of 17q21 variants and smoking exposure in early-onset asthma. N. Engl. J. Med. 359, 1985–1994 (2008).
Galanter, J. et al. ORMDL3 gene is associated with asthma in three ethnically diverse populations. Am. J. Respir. Crit. Care Med. 177, 1194–1200 (2008).
Halapi, E. et al. A sequence variant on 17q21 is associated with age at onset and severity of asthma. Eur. J. Hum. Genet. 18, 902–908 (2010).
Leung, T.F. et al. Asthma and atopy are associated with chromosome 17q21 markers in Chinese children. Allergy 64, 621–628 (2009).
Madore, A.M., Tremblay, K., Hudson, T.J. & Laprise, C. Replication of an association between 17q21 SNPs and asthma in a French-Canadian familial collection. Hum. Genet. 123, 93–95 (2008).
Sleiman, P.M. et al. ORMDL3 variants associated with asthma susceptibility in North Americans of European ancestry. J. Allergy Clin. Immunol. 122, 1225–1227 (2008).
Tavendale, R., Macgregor, D.F., Mukhopadhyay, S. & Palmer, C.N. A polymorphism controlling ORMDL3 expression is associated with asthma that is poorly controlled by current medications. J. Allergy Clin. Immunol. 121, 860–863 (2008).
Wu, H. et al. Genetic variation in ORM1-like 3 (ORMDL3) and gasdermin-like (GSDML) and childhood asthma. Allergy 64, 629–635 (2009).
Bisgaard, H. et al. Chromosome 17q21 gene variants are associated with asthma and exacerbations but not atopy in early childhood. Am. J. Respir. Crit. Care Med. 179, 179–185 (2009).
Moffatt, M.F. et al. A large-scale, consortium-based genomewide association study of asthma. N. Engl. J. Med. 363, 1211–1221 (2010).
Choubey, D. et al. Interferon-inducible p200-family proteins as novel sensors of cytoplasmic DNA: role in inflammation and autoimmunity. J. Interferon Cytokine Res. 30, 371–380 (2010).
Li, Y., Willer, C.J., Ding, J., Scheet, P. & Abecasis, G.R. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol. 34, 816–834 (2010).
Howie, B.N., Donnelly, P. & Marchini, J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 5, e1000529 (2009).
Acknowledgements
This work was supported by grants from the Office of the Director, NIH to C.O. and D.L.N. and the National Heart, Lung, and Blood Institute (HL101651 to C.O. and D.L.N.; HL087665 to D.L.N.; HL070831, HL072414 and HL049596 to C.O.; HL064307 and HL064313 to F.D.M.; HL075419, HL65899, HL083069, HL066289, HL087680, HL101543 and HL101651 to S.T.W.; HL079055 to L.K.W.; HL087699, HL49612, HL075417, HL04266 and HL072433 to K.C.B.; HL061768 and HL076647 to F.D.G.; HL087680 to W.J.G.; HL078885 and HL088133 to E.G.B.; HL87665 to D.A.M.; and HL69116, HL69130, HL69149, HL69155, HL69167, HL69170, HL69174 and HL69349 to D.A.M., E.R.B., W.W.B., W.J.C., M.C., K.F.C., S.C.E., E.I. and S.E.W.), the National Institutes of Allergy and Infectious Disease (AI070503 to C.O.; AI079139 and AI061774 to L.K.W.; AI50024, AI44840 and AI41040 to K.C.B.; and AI077439 to E.G.B.), the National Institute of Diabetes and Digestive and Kidney Diseases to L.K.W. (DK064695); the National Institutes of Environmental Health Sciences (ES09606, ES018176 and ES015903 to K.C.B.; ES007048, ES009581, R826708, RD831861 and ES011627 to F.D.G.; ES015794 to E.G.B.; and the Division of Intramural Research, Z01 ES049019, to S.J.L.); the National Center for Research Resources (RR03048 to K.C.B.), the Environmental Protection Agency (83213901 and R-826724 to K.C.B.), the American Asthma Foundation and the Fund for Henry Ford Hospital (to L.K.W.), Mary Beryl Patch Turnbull Scholar Program (to K.C.B.), the National Council of Science and Technology (Mexico) (26206-M to I. Ruczinski), the Centers for Disease Control, US (to I. Ruczinski), the Eudowood Foundation (to N.N.H.); and the Flight Attendant Medical Research Institute (FAMRI), Robert Wood Johnson Foundation (RWJF) Amos Medical Faculty Development Award, the American Asthma Foundation, and the Sandler Foundation (to E.G.B.).
The authors gratefully acknowledge the contributions of H. Moreno-Macias, A. Barraza-Villarreal and M. Ramirez-Aguilar to the MCCAS; R. McConnell, D.C. Thomas, E. Avol, K. Berhane, J. Liu, E. Rappaport and C. Edlund to the CHS; L.N. Borrell, H. Farber, R. Kumar and D. Serebrisky to the GALA and GALA2 studies; S. Thyne to the GALA2 and SAGE studies; M. LeNoir, K. Meade and H. Geoff Watson to the SAGE study; and A.V. Grant, L. Gao, C. Vergara, Y.J. Tsai, P. Gao, M.C. Liu, P. Breysse, M.B. Bracken, J. Hoh, E.W. Pugh, A.F. Scott, G. Abecasis, T. Murray, T. Hand, M. Yang, M. Campbell, C. Foster, J.B. Hetmanski, R. Ashworth, C.M. Ongaco, K.N. Hetrick and K.F. Doheny to the GRAAD study. The authors also acknowledge the support from J. Kiley, S. Banks-Schlegel and W. Gan at the National Heart, Lung, and Blood Institute and all the subjects and families that participated in these studies.
Author information
Authors and Affiliations
Consortia
Contributions
E.R.B., B.A.R., D.A.M., S.J.L., F.D.G., W.J.G., E.G.B., F.D.M., S.T.W., L.K.W., K.C.B., C.O. and D.L.N. conceived and designed the experiments. W.J.G., A.M.L., C.E., D.J.V.D.B., M.T.S., K.D.R., E.R.B., D.A.M., S.T.W., E.G.B. and L.K.W. performed the experiments. D.G.T., E.J.A., G.Y.C., W.J.G., C.R.G., B.E.H., A.M.L., R.A. Mathias, D.B.H., J.W.B., D.A.S., N.R., D.C., D.V.C., L.A.R., Xingnan L., D.H., R.D.H., J.G., M.S., V.J.M., L.L., I. Ruczinski, S.H., H.V., Xia L., T.H.B., E.R.B., D.A.M. and D.L.N. performed statistical analysis. D.G.T., E.J.A., G.Y.C., W.J.G., C.R.G., B.E.H., A.M.L., R.A. Mathias, D.B.H., J.W.B., C.E., D.A.S., N.R., D.C., D.V.C., L.A.R., Xingnan L., R.A. Myers, D.H., M.T.S., J.G., V.J.M., L.L., I. Romieu, S.H., H.V., Xia L., T.H.B., E.R.B., D.A.M., F.D.G., L.K.W., K.C.B. and D.L.N. analyzed the data. P.E.G., A.M.L., J.C.C., D.V.C., M.S.-Q., A.T., I. Romieu, N.N.H., E.I., M.T.S., J.G., P.C.A., L.A., J.R.R.-S., R.C., W.R.-C., G.B.D., N.F.A., M.S., M.U.F., G.M.D., H.R.W., S.C.E., J.G.F., K.F.C., W.J.C., M.C., J.-J.S.-M., B.d.R.-N., K.A.D., A.H., S.E.W., W.W.B., J.E.G., R.F.L., E.R.B., D.A.M., S.J.L., F.D.G., F.D.M., S.T.W., L.K.W., E.G.B., K.C.B. and C.O. contributed reagents, materials and analysis tools. D.G.T., G.Y.C., W.J.G., B.E.H., A.M.L., R.A. Mathias, D.B.H., E.R.B., B.A.R., D.A.M., S.J.L., F.D.G., E.G.B., F.D.M., S.T.W., L.K.W., K.C.B., C.O. and D.L.N. wrote the paper. R.A.A. coordinated the study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–16, Supplementary Tables 1–8 and Supplementary Note. (PDF 4161 kb)
Rights and permissions
About this article
Cite this article
Mexico City Childhood Asthma Study (MCAAS)., Children's Health Study (CHS) and HARBORS study., Genetics of Asthma in Latino Americans (GALA) Study, the Study of Genes-Environment and Admixture in Latino Americans (GALA2) and the Study of African Americans, Asthma, Genes & Environments (SAGE). et al. Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nat Genet 43, 887–892 (2011). https://doi.org/10.1038/ng.888
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.888
This article is cited by
-
Association between interleukin-6-174G/C gene polymorphism and asthma severity: exploring the role of total serum IgE, blood eosinophils, and FeNO as markers of type 2 inflammation
Allergy, Asthma & Clinical Immunology (2024)
-
Bronchial epithelial transcriptomics and experimental validation reveal asthma severity-related neutrophilc signatures and potential treatments
Communications Biology (2024)
-
Role of thymic stromal lymphopoietin in allergy and beyond
Nature Reviews Immunology (2023)
-
Inflammatory and infectious upper respiratory diseases associate with 41 genomic loci and type 2 inflammation
Nature Communications (2023)
-
Exploring the significance of interleukin-33/ST2 axis in minimal change disease
Scientific Reports (2023)