Abstract
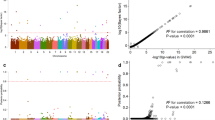
To identify susceptibility loci for meningioma, we conducted a genome-wide association study of 859 affected individuals (cases) and 704 controls with validation in two independent sample sets totaling 774 cases and 1,764 controls. We identified a new susceptibility locus for meningioma at 10p12.31 (MLLT10, rs11012732, odds ratio = 1.46, Pcombined = 1.88 × 10−14). This finding advances our understanding of the genetic basis of meningioma development.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Mawrin, C. & Perry, A . J. Neurooncol. 99, 379–391 (2010).
Simon, M., Bostrom, J.P. & Hartmann, C. Neurosurgery 60, 787–798 (2007).
Wiemels, J., Wrensch, M. & Claus, E.B. J. Neurooncol. 99, 307–314 (2010).
Hemminki, K. & Li, X. Cancer Epidemiol. Biomarkers Prev. 12, 1137–1142 (2003).
Clayton, D.G. et al. Nat. Genet. 37, 1243–1246 (2005).
Price, A.L. et al. Nat. Genet. 38, 904–909 (2006).
Mihaila, D. et al. Clin. Cancer Res. 9, 4443–4451 (2003).
Mahmoudi, T. et al. PLoS Biol. 8, e1000539 (2010).
Saydam, O. et al. Mol. Cell. Biol. 29, 5923–5940 (2009).
Matys, V. et al. Nucleic Acids Res. 34, D108–D110 (2006).
Ferretti, V. et al. Nucleic Acids Res. 35, D122–D126 (2007).
Stranger, B.E. et al. PLoS Genet. 1, e78 (2005).
Stranger, B.E. et al. Science 315, 848–853 (2007).
Hepworth, S.J. et al. Br. Med. J. 332, 883–887 (2006).
Acknowledgements
Cancer Research UK (C1298/A8362 supported by the Bobby Moore Fund) provided principal funding for the study. Additional support for the UK study was provided by the DJ Fielding Medical Research Trust. We acknowledge National Health Service (NHS) funding to the National Institute for Health Research (NIHR) Biomedical Research Centre.
The UK, Swedish and Danish INTERPHONE studies were supported by the European Union Fifth Framework Program 'Quality of Life and Management of Living Resources' (contract number QLK4-CT-1999-01563) and the International Union against Cancer (UICC). The UICC received funds for this purpose from the Mobile Manufacturers' Forum and Global System for Mobile Communications Association. Provision of funds from the UICC was governed by agreements that guaranteed INTERPHONE's complete scientific independence. These agreements are publicly available at http://www.iarc.fr/en/research-groups/RAD/RCAd.html. The Swedish INTERPHONE center was also supported by the Swedish Research Council, the Cancer Foundation of Northern Sweden, the Swedish Cancer Society and the Nordic Cancer Union, and the Danish center was supported by the Danish Cancer Society. The UK centers were also supported by the Mobile Telecommunications and Health Research (MTHR) Programme, and the Northern UK center was supported by the Health and Safety Executive, Department of Health and Safety Executive and the UK Network Operators (O2, Orange, T-Mobile, Vodafone and '3'). A.A. received support from the Emil Aaltonen Foundation. We acknowledge the participation of the clinicians and other hospital staff, cancer registries and study staff who contributed to the blood sample and data collection for this study (as detailed in ref. 14).
In Germany, funding was provided to M.S. and J.S. by the Deutsche Forschungsgemeinschaft (Si 552, Schr 285), the Deutsche Krebshilfe (70-2385-Wi2, 70-3163-Wi3, 10-6262) and BONFOR. We are indebted to R. Mahlberg (Bonn), who provided technical support.
The GWAS made use of genotyping data from the population-based HNR study. The HNR study is supported by the Heinz Nixdorf Foundation (Germany). Additionally, the study is funded by the German Ministry of Education and Science and the German Research Council (DFG; Project SI 236/8-1, SI236/9-1, ER 155/6-1). Funding was provided to L.E. by the Medical Faculty of the University Hospital of Essen (IFORES). The genotyping of the Illumina HumanOmni-1 Quad BeadChips of the HNR subjects was financed by the German Centre for Neurodegenerative Disorders (DZNE), Bonn. We are extremely grateful to all investigators who contributed to the generation of this dataset.
Finally, we are grateful to all subjects and individuals for their participation, and we would also like to thank the clinicians and other hospital staff, cancer registries and study staff in respective centers who contributed to the blood sample and data collection.
Author information
Authors and Affiliations
Contributions
R.S.H. and M.S. conceived the study. R.S.H. designed the study and obtained financial support. R.S.H., M.S. and S.E.D. drafted the manuscript. S.E.D., P.B., F.J.H. and Y.P.M. performed statistical and bioinformatic analyses. P.B. managed sample coordination and laboratory analyses. B.O. and A.L. performed genotyping. M.S. and J.S. collected the German (Bonn) cases and obtained funding. M.S. oversaw sample preparation in Bonn. In the UK, A.S., M.J.S., K.M., S.J.H. and R.S.H. developed subject recruitment, sample acquisition and performed sample collection of cases. S.M. and L.E. managed collection of HNR controls. P.H. and T.W.M. performed genotyping of the HNR controls. M.M.N. managed sample coordination and laboratory analyses. For the Swedish INTERPHONE study, M.F., S.L. and A.A. developed the study design and conducted subject recruitment and control selection. M.F., S.L., A.A., B.M. and R.H. organized sample acquisition and performed sample collection of case and controls. U.A. coordinated sample collection and complied information into data files of cases and controls for statistical analyses, and B.M. and R.H. performed laboratory management and oversaw DNA extraction. The NSHDS samples were collected by Umeå University (principal investigator G. Hallmans), and the additional samples were collected at the neurosurgery department in Umeå from 2005 and onwards (T.B. and R.H.) and through the national GLIOGENE study (principal investigator B.M.). In Denmark, C.J. and M.K. conducted subject recruitment and sample collection. All authors contributed to the final paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Methods, Supplementary Tables 1–3 and Supplementary Figures 1–4. (PDF 3003 kb)
Rights and permissions
About this article
Cite this article
Dobbins, S., Broderick, P., Melin, B. et al. Common variation at 10p12.31 near MLLT10 influences meningioma risk. Nat Genet 43, 825–827 (2011). https://doi.org/10.1038/ng.879
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.879