Abstract
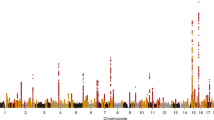
We carried out a genome-wide association study of IgA nephropathy, a major cause of kidney failure worldwide. We studied 1,194 cases and 902 controls of Chinese Han ancestry, with targeted follow up in Chinese and European cohorts comprising 1,950 cases and 1,920 controls. We identified three independent loci in the major histocompatibility complex, as well as a common deletion of CFHR1 and CFHR3 at chromosome 1q32 and a locus at chromosome 22q12 that each surpassed genome-wide significance (P values for association between 1.59 × 10−26 and 4.84 × 10−9 and minor allele odds ratios of 0.63–0.80). These five loci explain 4–7% of the disease variance and up to a tenfold variation in interindividual risk. Many of the alleles that protect against IgA nephropathy impart increased risk for other autoimmune or infectious diseases, and IgA nephropathy risk allele frequencies closely parallel the variation in disease prevalence among Asian, European and African populations, suggesting complex selective pressures.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Coresh, J. et al. Prevalence of chronic kidney disease in the United States. J. Am. Med. Assoc. 298, 2038–2047 (2007).
Tsukamoto, Y. et al. Report of the Asian Forum of Chronic Kidney Disease Initiative (AFCKDI) 2007. “Current status and perspective of CKD in Asia”: diversity and specificity among Asian countries. Clin. Exp. Nephrol. 13, 249–256 (2009).
Gesualdo, L., Di Palma, A.M., Morrone, L.F., Strippoli, G.F. & Schena, F.P. The Italian experience of the national registry of renal biopsies. Kidney Int. 66, 890–894 (2004).
D'Amico, G. The commonest glomerulonephritis in the world: IgA nephropathy. Q. J. Med. 64, 709–727 (1987).
Nair, R. & Walker, P.D. Is IgA nephropathy the commonest primary glomerulopathy among young adults in the USA? Kidney Int. 69, 1455–1458 (2006).
Varis, J. et al. Immunoglobulin and complement deposition in glomeruli of 756 subjects who had committed suicide or met with a violent death. J. Clin. Pathol. 46, 607–610 (1993).
Suzuki, K. et al. Incidence of latent mesangial IgA deposition in renal allograft donors in Japan. Kidney Int. 63, 2286–2294 (2003).
Kiryluk, K. et al. Genetic studies of IgA nephropathy: past, present, and future. Pediatr. Nephrol. 25, 2257–2268 (2010).
Barratt, J. & Feehally, J. IgA nephropathy. J. Am. Soc. Nephrol. 16, 2088–2097 (2005).
Hastings, M.C. et al. Galactose-deficient IgA1 in African Americans with IgA nephropathy: serum levels and heritability. Clin. J. Am. Soc. Nephrol. 5, 2069–2074 (2010).
Gharavi, A.G. et al. Aberrant IgA1 glycosylation is inherited in familial and sporadic IgA nephropathy. J. Am. Soc. Nephrol. 19, 1008–1014 (2008).
Lin, X. et al. Aberrant galactosylation of IgA1 is involved in the genetic susceptibility of Chinese patients with IgA nephropathy. Nephrol. Dial. Transplant. 24, 3372–3375 (2009).
Moldoveanu, Z. et al. Patients with IgA nephropathy have increased serum galactose-deficient IgA1 levels. Kidney Int. 71, 1148–1154 (2007).
Mestecky, J. et al. Defective galactosylation and clearance of IgA1 molecules as a possible etiopathogenic factor in IgA nephropathy. Contrib. Nephrol. 104, 172–182 (1993).
Tomana, M. et al. Circulating immune complexes in IgA nephropathy consist of IgA1 with galactose-deficient hinge region and antiglycan antibodies. J. Clin. Invest. 104, 73–81 (1999).
Gharavi, A.G. et al. IgA nephropathy, the most common cause of glomerulonephritis, is linked to 6q22–23. Nat. Genet. 26, 354–357 (2000).
Bisceglia, L. et al. Genetic heterogeneity in Italian families with IgA nephropathy: suggestive linkage for two novel IgA nephropathy loci. Am. J. Hum. Genet. 79, 1130–1134 (2006).
Paterson, A.D. et al. Genome-wide linkage scan of a large family with IgA nephropathy localizes a novel susceptibility locus to chromosome 2q36. J. Am. Soc. Nephrol. 18, 2408–2415 (2007).
Feehally, J. et al. HLA has strongest association with IgA nephropathy in genome-wide analysis. J. Am. Soc. Nephrol. 21, 1791–1797 (2010).
Storey, J.D. & Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 100, 9440–9445 (2003).
de Bakker, P.I. et al. A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat. Genet. 38, 1166–1172 (2006).
Hughes, A.E. et al. A common CFH haplotype, with deletion of CFHR1 and CFHR3, is associated with lower risk of age-related macular degeneration. Nat. Genet. 38, 1173–1177 (2006).
Raychaudhuri, S. et al. Associations of CFHR1-CFHR3 deletion and a CFH SNP to age-related macular degeneration are not independent. Nat. Genet. 42, 553–555, author reply 555–556 (2010).
Davila, S. et al. Genome-wide association study identifies variants in the CFH region associated with host susceptibility to meningococcal disease. Nat. Genet. 42, 772–776 (2010).
Barcellos, L.F. et al. High-density SNP screening of the major histocompatibility complex in systemic lupus erythematosus demonstrates strong evidence for independent susceptibility regions. PLoS Genet. 5, e1000696 (2009).
Erlich, H. et al. HLA DR-DQ haplotypes and genotypes and type 1 diabetes risk: analysis of the type 1 diabetes genetics consortium families. Diabetes 57, 1084–1092 (2008).
Ferreira, R.C. et al. Association of IFIH1 and other autoimmunity risk alleles with selective IgA deficiency. Nat. Genet. 42, 777–780 (2010).
Imielinski, M. et al. Common variants at five new loci associated with early-onset inflammatory bowel disease. Nat. Genet. 41, 1335–1340 (2009).
Kamatani, Y. et al. A genome-wide association study identifies variants in the HLA-DP locus associated with chronic hepatitis B in Asians. Nat. Genet. 41, 591–595 (2009).
Singer, J.B. et al. A genome-wide study identifies HLA alleles associated with lumiracoxib-related liver injury. Nat. Genet. 42, 711–714 (2010).
Zhou, X. et al. HLA-DPB1 and DPB2 are genetic loci for systemic sclerosis: a genome-wide association study in Koreans with replication in North Americans. Arthritis Rheum. 60, 3807–3814 (2009).
Mignot, E. et al. Complex HLA-DR and -DQ interactions confer risk of narcolepsy-cataplexy in three ethnic groups. Am. J. Hum. Genet. 68, 686–699 (2001).
Begley, G.S., Horvath, A.R., Taylor, J.C. & Higgins, C.F. Cytoplasmic domains of the transporter associated with antigen processing and P-glycoprotein interact with subunits of the proteasome. Mol. Immunol. 42, 137–141 (2005).
Muchamuel, T. et al. A selective inhibitor of the immunoproteasome subunit LMP7 blocks cytokine production and attenuates progression of experimental arthritis. Nat. Med. 15, 781–787 (2009).
Coppo, R. et al. Upregulation of the immunoproteasome in peripheral blood mononuclear cells of patients with IgA nephropathy. Kidney Int. 75, 536–541 (2009).
Atkinson, J.P. & Goodship, T.H. Complement factor H and the hemolytic uremic syndrome. J. Exp. Med. 204, 1245–1248 (2007).
Heinen, S. et al. Factor H-related protein 1 (CFHR-1) inhibits complement C5 convertase activity and terminal complex formation. Blood 114, 2439–2447 (2009).
Esashi, E. et al. Oncostatin M deficiency leads to thymic hypoplasia, accumulation of apoptotic thymocytes and glomerulonephritis. Eur. J. Immunol. 39, 1664–1670 (2009).
Wojtasz, L. et al. Mouse HORMAD1 and HORMAD2, two conserved meiotic chromosomal proteins, are depleted from synapsed chromosome axes with the help of TRIP13 AAA-ATPase. PLoS Genet. 5, e1000702 (2009).
Grossman, S.R. et al. A composite of multiple signals distinguishes causal variants in regions of positive selection. Science 327, 883–886 (2010).
Maller, J. et al. Common variation in three genes, including a noncoding variant in CFH, strongly influences risk of age-related macular degeneration. Nat. Genet. 38, 1055–1059 (2006).
Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447, 661–678 (2007).
Zipfel, P.F. et al. Deletion of complement factor H-related genes CFHR1 and CFHR3 is associated with atypical hemolytic uremic syndrome. PLoS Genet. 3, e41 (2007).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Skol, A.D., Scott, L.J., Abecasis, G.R. & Boehnke, M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat. Genet. 38, 209–213 (2006).
Clayton, D. & Leung, H.T. An R package for analysis of whole-genome association studies. Hum. Hered. 64, 45–51 (2007).
Browning, S.R. & Browning, B.L. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 81, 1084–1097 (2007).
Gusev, A. et al. Whole population, genome-wide mapping of hidden relatedness. Genome Res. 19, 318–326 (2009).
Conrad, D.F. et al. Origins and functional impact of copy number variation in the human genome. Nature 464, 704–712 (2010).
Craddock, N. et al. Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature 464, 713–720 (2010).
Acknowledgements
We are grateful to all study participants for their contribution to this work. We also thank the staff of the Yale West Campus Center for Genome Analysis for their excellent support. We also appreciate the assistance of C.V. Barker and S.Y. Woodford with sample collection. This study was supported by RC1DK087445 (A.G.G., R.P.L.), R01DK082753 (A.G.G., J.N., B.A.J. and R.J.W.) and KL2 RR24157 (K.K.), the Center for Glomerular Diseases at Columbia University, the Yale Center Translational Science Award and the Yale Center for Human Genetics and Genomics. R.P.L. is an investigator of the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
Subject clinical characterization, recruitment and contribution of samples: P.H., J.X., S.S.C., B.A.J., R.J.W., J.N., J.C.H., H.W., J.L., L.Z., W.W., Z.W., S.S., R. Magistroni, G.M.G., M.B., P.R., C.P., L.A., G.B., G.F., A. Amore, L.P., R.C., C.I., B.F.V., E.P., M.S., R. Mignani, L.G., F.B., P.M., A. Amoroso, F.S., N.C. and H.Z.
DNA preparation: Y.L., P.H., J.X., F.L., I.B., K.K., C.J.M. and M.C.
Genotyping and wet lab experiments: S.M., S.U., I.T., C.J.M., M.C., P.H., J.X. and Y.L.
Data management: K.K., Y.L., S.S.C. and M.C.
Data analysis: K.K., M.C., A.G.G. and R.P.L.
Analytical support and discussion: K.Y. and M.G.
Manuscript preparation: A.G.G., K.K., M.C. and R.P.L.
Conception and overall supervision of project: A.G.G. and R.P.L.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Note, Supplementary Tables 1–17 and Supplementary Figures 1–8. (PDF 2083 kb)
Rights and permissions
About this article
Cite this article
Gharavi, A., Kiryluk, K., Choi, M. et al. Genome-wide association study identifies susceptibility loci for IgA nephropathy. Nat Genet 43, 321–327 (2011). https://doi.org/10.1038/ng.787
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.787
This article is cited by
-
Genetics of IgA nephrology: risks, mechanisms, and therapeutic targets
Pediatric Nephrology (2024)
-
Complement inhibitors in pediatric kidney diseases: new therapeutic opportunities
Pediatric Nephrology (2024)
-
GWAS for the composite traits of hematuria and albuminuria
Scientific Reports (2023)
-
IgA nephropathy
Nature Reviews Disease Primers (2023)
-
Diagnostik und Therapie IgA Nephropathie – 2023
Wiener klinische Wochenschrift (2023)