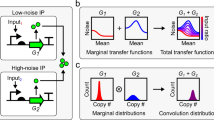
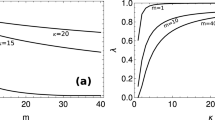
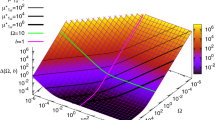
Abstract
Gene expression involves inherently probabilistic steps that create fluctuations in protein abundances. The results from many in-depth analyses and genome-scale surveys have suggested how such fluctuations arise and spread, often in ways consistent with stochastic models of transcription and translation. But fluctuations also arise during cell division when molecules are partitioned stochastically between the two daughters. Here we mathematically demonstrate how stochastic partitioning contributes to the non-genetic heterogeneity. Our results show that partitioning errors are hard to correct, and that the resulting noise profiles are remarkably difficult to separate from gene expression noise. By applying these results to common experimental strategies and distinguishing between creation versus transmission of noise, we hypothesize that much of the cell-to-cell heterogeneity that has been attributed to various aspects of gene expression instead comes from random segregation at cell division. We propose experiments to separate between these two types of fluctuations and discuss future directions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Warren, G. Membrane partitioning during cell division. Annu. Rev. Biochem. 62, 323–348 (1993).
Marshall, W.F. Stability and robustness of an organelle number control system: modeling and measuring homeostatic regulation of centriole abundance. Biophys. J. 93, 1818–1833 (2007).
Birky, C.W. Jr. The partitioning of cytoplasmic organelles at cell division. Int. Rev. Cytol. Suppl. 15, 49–89 (1983).
Nordströem, K. & Austin, S.J. Mechanisms that contribute to the stable segregation of plasmids. Annu. Rev. Genet. 23, 37–69 (1989).
Mantzaris, N.V. From single-cell genetic architecture to cell population dynamics: quantitatively decomposing the effects of different population heterogeneity sources for a genetic network with positive feedback architecture. Biophys. J. 92, 4271–4288 (2007).
Cookson, N.A., Cookson, S.W., Tsimring, L.S. & Hasty, J. Cell cycle-dependent variations in protein concentration. Nucleic Acids Res. 38, 2676–2681 (2010).
Blake, W.J., Kaern, M., Cantor, C.R. & Collins, J.J. Noise in eukaryotic gene expression. Nature 422, 633–637 (2003).
Raser, J.M. & O′Shea, E.K. Control of stochasticity in eukaryotic gene expression. Science 304, 1811–1845 (2004).
Bar-Even, A. et al. Noise in protein expression scales with natural protein abundance. Nat. Genet. 38, 636–643 (2006).
Taniguchi, Y. et al. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science 329, 533–538 (2010).
Newman, J.R. et al. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature 441, 840–846 (2006).
Zenklusen, D., Larson, D.R. & Singer, R.H. Single-RNA counting reveals alternative modes of gene expression in yeast. Nat. Struct. Mol. Biol. 15, 1263–1271 (2008).
Ozbudak, E.M., Thattai, M., Kurtser, I., Grossman, A.D. & van Oudenaarden, A. Regulation of noise in the expression of a single gene. Nat. Genet. 31, 69–73 (2002).
Elowitz, M.B., Levine, A.J., Siggia, E.D. & Swain, P.S. Stochastic gene expression in a single cell. Science 297, 1183–1186 (2002).
Paulsson, J. & Ehrenberg, M. Noise in a minimal regulatory network: plasmid copy number control. Q. Rev. Biophys. 34, 1–59 (2001).
Paulsson, J. Summing up the noise in gene networks. Nature 427, 415–418 (2004).
Paulsson, J. Models of stochastic gene expression. Phys. Life Rev. 2, 157–175 (2005).
Berg, O.G. A model for the statistical fluctuations of protein numbers in a microbial population. J. Theor. Biol. 71, 587–603 (1978).
Rigney, D.R. Stochastic model of constitutive protein levels in growing and dividing bacterial cells. J. Theor. Biol. 76, 453–480 (1979).
Swain, P.S., Elowitz, M.B. & Siggia, E.D. Intrinsic and extrinsic contributions to stochasticity in gene expression. Proc. Natl. Acad. Sci. USA 99, 12795–12800 (2002).
Golding, I., Paulsson, J., Zawilski, S.M. & Cox, E.C. Real-time kinetics of gene activity in individual bacteria. Cell 123, 1025–1036 (2005).
Chubb, J.R., Trcek, T., Shenoy, S.M. & Singer, R.H. Transcriptional pulsing of a developmental gene. Curr. Biol. 16, 1018–1025 (2006).
Pedraza, J.M. & Paulsson, J. Effects of molecular memory and bursting on fluctuations in gene expression. Science 319, 339–343 (2008).
Paulsson, J. & Ehrenberg, M. Random signal fluctuations can reduce random fluctuations in regulated components of chemical regulatory networks. Phys. Rev. Lett. 84, 5447–5450 (2000).
Lestas, I., Vinnicombe, G. & Paulsson, J. Fundamental limits on the suppression of molecular fluctuations. Nature 467, 174–178 (2010).
Maxwell, J.C. On governors. Proceedings of the Royal Society 16, 270–283 (1867).
Cannon, W.B. The Wisdom of the Body (Norton, New York, New York, USA,1932).
Kar, S., Baumann, W.T., Paul, M.R. & Tyson, J.J. Exploring the roles of noise in the eukaryotic cell cycle. Proc. Natl. Acad. Sci. USA 106, 6471–6476 (2009).
Palsson, B.O. & Lightfoot, E.N. Mathematical modelling of dynamics and control in metabolic networks. V. Static bifurcations in single biochemical control loops. J. Theor. Biol. 113, 279–298 (1985).
Bergeland, T., Widerberg, J., Bakke, O. & Nordeng, T.W. Mitotic partitioning of endosomes and lysosomes. Curr. Biol. 11, 644–651 (2001).
McAuley, P.J. Partitioning of symbiotic chlorella at host cell telophase in the green hydra symbiosis. Phil. Trans. R. Soc. Lond. B 329, 47–53 (1990).
Savage, D.F., Afonso, B., Chen, A.H. & Silver, P.A. Spatially ordered dynamics of the bacterial carbon fixation machinery. Science 327, 1258–1261 (2010).
Wilson, E.B. The distribution of the chondriosomes to the spermatozoa in scorpions. Proc. Natl. Acad. Sci. USA 2, 321–324 (1916).
Wilson, E.B. The distribution of sperm-forming materials in scorpions. J. Morphol. Physiol. 52, 429–483 (1931).
Hood, R.D., Watson, O.F., Deason, T.R. & Benton, C.L.B. Jr. Ultrastructure of scorpion spermatozoa with atypical axonemes. Cytobios 5, 167–177 (1972).
Shima, D.T., Haldar, K., Pepperkok, R., Watson, R. & Warren, G. Partitioning of the Golgi apparatus during mitosis in living HeLa cells. J. Cell Biol. 137, 1211–1228 (1997).
Hennis, A.S. & Birky, C.W. Stochastic partitioning of chloroplasts at cell division in the alga Olisthodiscus, and compensating control of chloroplast replication. J. Cell Sci. 70, 1–15 (1984).
Sheahan, M.B., Rose, R.J. & McCurdy, D.W. Organelle inheritance in plant cell division: the actin cytoskeleton is required for unbiased inheritance of chloroplasts, mitochondria and endoplasmic reticulum in dividing protoplasts. Plant J. 37, 379–390 (2004).
Rosenfeld, N., Young, J.W., Alon, U., Swain, P.S. & Elowitz, M.B. Gene regulation at the single-cell level. Science 307, 1962–1965 (2005).
Yu, J., Xiao, J., Ren, X., Lao, K. & Xie, X.S. Probing gene expression in live cells, one protein molecule at a time. Science 311, 1600–1603 (2006).
Cai, L., Friedman, N. & Xie, X.S. Stochastic protein expression in individual cells at the single molecule level. Nature 440, 358–362 (2006).
Raj, A., Peskin, C.S., Tranchina, D., Vargas, D.Y. & Tyagi, S. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 4, e309 (2006).
Grünwald, D., Singer, R.H., Czaplinski, K. & Lynne, E.M.M.K. Chapter 27 Cell biology of mRNA decay. in Methods in Enzymology 448, 553–577 (Academic Press, 2008).
Paulsson, J. Prime movers of noisy gene expression. Nat. Genet. 37, 925–926 (2005).
Paulsson, J., Berg, O.G. & Ehrenberg, M. Stochastic focusing: fluctuation-enhanced sensitivity of intracellular regulation. Proc. Natl. Acad. Sci. USA 97, 7148–7153 (2000).
Acknowledgements
The research was supported by the National Science Foundation grants DMS-074876-0 and CAREER 0720056 and by grant GM081563-02 from the US National Institutes of Health. We thank I. Golding for valuable comments on the manuscript.
Author information
Authors and Affiliations
Contributions
D.H. and J.P. jointly conceived the study, derived the results and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Huh, D., Paulsson, J. Non-genetic heterogeneity from stochastic partitioning at cell division. Nat Genet 43, 95–100 (2011). https://doi.org/10.1038/ng.729
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.729