Abstract
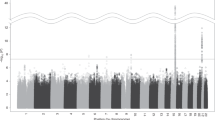
Consistent but indirect evidence has implicated genetic factors in smoking behavior1,2. We report meta-analyses of several smoking phenotypes within cohorts of the Tobacco and Genetics Consortium (n = 74,053). We also partnered with the European Network of Genetic and Genomic Epidemiology (ENGAGE) and Oxford-GlaxoSmithKline (Ox-GSK) consortia to follow up the 15 most significant regions (n > 140,000). We identified three loci associated with number of cigarettes smoked per day. The strongest association was a synonymous 15q25 SNP in the nicotinic receptor gene CHRNA3 (rs1051730[A], β = 1.03, standard error (s.e.) = 0.053, P = 2.8 × 10−73). Two 10q25 SNPs (rs1329650[G], β = 0.367, s.e. = 0.059, P = 5.7 × 10−10; and rs1028936[A], β = 0.446, s.e. = 0.074, P = 1.3 × 10−9) and one 9q13 SNP in EGLN2 (rs3733829[G], β = 0.333, s.e. = 0.058, P = 1.0 × 10−8) also exceeded genome-wide significance for cigarettes per day. For smoking initiation, eight SNPs exceeded genome-wide significance, with the strongest association at a nonsynonymous SNP in BDNF on chromosome 11 (rs6265[C], odds ratio (OR) = 1.06, 95% confidence interval (Cl) 1.04–1.08, P = 1.8 × 10−8). One SNP located near DBH on chromosome 9 (rs3025343[G], OR = 1.12, 95% Cl 1.08–1.18, P = 3.6 × 10−8) was significantly associated with smoking cessation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Rose, R.J., Broms, U., Korhonen, T., Dick, D.M. & Kaprio, J. Genetics of Smoking Behavior. in Handbook of Behavior Genetics, 1 (ed. Kim, Y.-K.) 411–432 (Springer, New York, 2009).
Li, M.D. Identifying susceptibility loci for nicotine dependence: 2008 update based on recent genome-wide linkage analyses. Hum. Genet. 123, 119–131 (2008).
Thorgeirsson, T.E. et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature 452, 638–642 (2008).
Fiore, M.C., Smith, S.S., Jorenby, D.E. & Baker, T.B. The effectiveness of the nicotine patch for smoking cessation. A meta-analysis. J. Am. Med. Assoc. 271, 1940–1947 (1994).
Li, Y., Willer, C., Sanna, S. & Abecasis, G. Genotype imputation. Annu. Rev. Genomics Hum. Genet. 10, 387–406 (2009).
de Bakker, P.I. et al. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum. Mol. Genet. 17, R122–R128 (2008).
Kraft, P., Zeggini, E. & Ioannidis, J.P.A. Replication in genome-wide association studies. Stat. Sci. published online, doi:10.1214/09-STS290 (2010).
Pereira, T.V., Patsopoulos, N.A., Salanti, G. & Ioannidis, J.P. Discovery properties of genome-wide association signals from cumulatively combined data sets. Am. J. Epidemiol. 170, 1197–1206 (2009).
Ioannidis, J.P., Patsopoulos, N.A. & Evangelou, E. Heterogeneity in meta-analyses of genome-wide association investigations. PLoS One 2, e841 (2007).
Pe'er, I. et al. Evaluating and improving power in whole-genome association studies using fixed marker sets. Nat. Genet. 38, 663–667 (2006).
Pe'er, I., Yelensky, R., Altshuler, D. & Daly, M.J. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet. Epidemiol. 32, 381–385 (2008).
Thorgeirsson, T. et al. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat. Genet. 42, 448–453 (2010).
Liu, J. et al. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat. Genet. 42, 436–440 (2010).
Saccone, N.L. et al. Multiple distinct risk loci for nicotine dependence identified by dense coverage of the complete family of nicotinic receptor subunit (CHRN) genes. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 150B, 453–466 (2009).
Nakajima, M. et al. Role of human cytochrome P4502A6 in C-oxidation of nicotine. Drug Metab. Dispos. 24, 1212–1217 (1996).
Mwenifumbo, J.C. & Tyndale, R.F. Molecular genetics of nicotine metabolism. Handb. Exp. Pharmacol. 192, 235–259 (2009).
Ray, R., Tyndale, R.F. & Lerman, C. Nicotine dependence pharmacogenetics: role of genetic variation in nicotine-metabolizing enzymes. J. Neurogenet. 23, 252–261 (2009).
Zhang, L.I. & Poo, M.M. Electrical activity and development of neural circuits. Nat. Neurosci. 4 Suppl, 1207–1214 (2001).
Levin, E.D., McClernon, F.J. & Rezvani, A.H. Nicotinic effects on cognitive function: behavioral characterization, pharmacological specification, and anatomic localization. Psychopharmacology (Berl.) 184, 523–539 (2006).
Gratacòs, M. et al. Brain-derived neurotrophic factor Val66Met and psychiatric disorders: meta-analysis of case-control studies confirm association to substance-related disorders, eating disorders, and schizophrenia. Biol. Psychiatry 61, 911–922 (2007).
Thorleifsson, G. et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat. Genet. 41, 18–24 (2009).
Zabetian, C.P. et al. A quantitative-trait analysis of human plasma-dopamine beta-hydroxylase activity: evidence for a major functional polymorphism at the DBH locus. Am. J. Hum. Genet. 68, 515–522 (2001).
Hindorff, L.A. et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. USA 106, 9362–9367 (2009).
Keskitalo, K. et al. Association of serum cotinine level with a cluster of three nicotinic acetylcholine receptor genes (CHRNA3/CHRNA5/CHRNB4) on chromosome 15. Hum. Mol. Genet. 18, 4007–4012 (2009).
Pomerleau, O.F. et al. Genetic research on complex behaviors: an examination of attempts to identify genes for smoking. Nicotine Tob. Res. 9, 883–901 (2007).
Lichtenstein, P. et al. The Swedish Twin Registry: a unique resource for clinical, epidemiological and genetic studies. J. Intern. Med. 252, 184–205 (2002).
Furberg, H., Lichtenstein, P., Pedersen, N.L., Bulik, C. & Sullivan, P.F. Cigarettes and oral snuff use in Sweden: prevalence and transitions. Addiction. 10, 1509–1515 (2006).
Kaprio, J., Pulkkinen, L. & Rose, R.J. Genetic and environmental factors in health-related behaviors: studies on Finnish twins and twin families. Twin Res. 5, 366–371 (2002).
Kaprio, J. & Koskenvuo, M. Genetic and environmental factors in complex diseases: the older Finnish Twin Cohort. Twin Res. 5, 358–365 (2002).
Centers for Disease Control and Prevention (CDC). Cigarette smoking among adults–United States, 2007. MMWR Morb. Mortal. Wkly. Rep. 57, 1221–1226 (2008); erratum 57, 1281 (2008).
Furberg, H., Lichtenstein, P., Pedersen, N.L., Bulik, C. & Sullivan, P.F. Cigarettes and oral snuff use in Sweden: Prevalence and transitions. Addiction 101, 1509–1515 (2006).
Frazer, K.A. et al. A second generation human haplotype map of over 3.1 million SNPs. Nature 449, 851–861 (2007).
Pritchard, J.K., Stephens, M. & Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 155, 945–959 (2000).
Price, A.L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006).
Li, Y., Ding, J. & Abecasis, G.R. MACH 1.0: rapid haplotype reconstruction and missing genotype inference. Am. J. Hum. Genet. S79, 2290 (2006).
Marchini, J., Howie, B., Myers, S., McVean, G. & Donnelly, P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 39, 906–913 (2007).
Servin, B. & Stephens, M. Imputation-based analysis of association studies: candidate regions and quantitative traits. PLoS Genet. 3, e114 (2007).
Lin, D.Y. & Zeng, D. Proper analysis of secondary phenotype data in case-control association studies. Genet. Epidemiol. 33, 256–265 (2009).
Devlin, B. & Roeder, K. Genomic control for association studies. Biometrics 55, 997–1004 (1999).
Centers for Disease Control and Prevention (CDC). Cigarette smoking among adults-United States, 2006. MMWR CDC Surveill. Summ. 56, 1157–1161 (2007).
Dudbridge, F. & Gusnanto, A. Estimation of significance thresholds for genomewide association scans. Genet. Epidemiol. 32, 227–234 (2008).
Acknowledgements
This work was funded by the University of North Carolina Lineberger Comprehensive Cancer Center University Cancer Research Fund Award and by US National Cancer Institute K07 CA118412 to H.F. Statistical analyses were carried out on the Genetic Cluster Computer (see URLs), which is supported by the Netherlands Scientific Organization (NWO 480-05-003). Acknowledgments for studies included in TAG are listed in the Supplementary Note.
Author information
Authors and Affiliations
Consortia
Contributions
TAG: study conception, design, management: H.F., P.F.S., Y.K., J. Dackor; TAG Statistical Working Group: D.-Y.L., P.K., J.P.A.I., D.P., H.F., Y.K., J. Dackor, S.P.F., N.F., E.H.L., J.D.M., J.M.V., D.I.B., D.L., B.M.E., E.L.T., B. McKnight, P.F.S., D. Absher; TAG Phenotype Working Group: C. Lerman, J.K., H.H.M., L.M.T., J.A.-M., E.H.L., J.E.R., M.D.L., J.M.V., H.F., Y.K., J. Dackor, S.P.F., P.F.S., E.L.T.; data analysis: Y.K., D.M.A., F.G., E.H.L., J.D.M., J.M.V., A.U.J., L. Bernardinelli, S.R.P., S.-J.H., B.M.E., C. Ladenvall, J.R.B.P., T.T., E.L.T., J.C.B., G.L., S.W.; TAG Manuscript Writing Group: H.F., Y.K., J. Dackor, P.F.S., C. Lerman, M.D.L., J.K., J.A.-M., P.K. All authors reviewed and approved the final version of the manuscript. The corresponding authors had access to the full data set of summary results contributed by each study.
ARIC: study conception, design, management: E.B.; phenotype collection, data management: N.F.; sample processing and genotyping: N.F.; data analysis: Y.K., N.F.
Atherosclerosis Thrombosis and Vascular Biology Italian Study Group: study conception, design, management: L. Bernardinelli, P.M.M., P.A.M., D. Ardissino; phenotype collection, data management: F.M., L. Bernandinelli; data analysis: L. Bernandinelli.
ADVANCE: study conception, design, management: S.P.F., D. Absher, T.Q., C.I., T.L.A., J.W.K.; phenotype collection, data management: S.P.F., T.Q., C.I., T.L.A., J.W.K.; sample processing and genotyping: D. Absher, T.Q.; data analysis: S.P.F., D. Absher, T.L.A., J.W.K.
Baltimore Longitudinal Study of Aging: study conception, design, management: L. Ferrucci; phenotype collection, data management: L. Ferrucci; data analysis: T.T.
CHS: study conception, design, management: B.M.P., J.C.B., C.D.F.; phenotype collection, data management: B.M.P.; sample processing and genotyping: T.H., K.D.T.; data analysis: B.M.P., E.L.T., J.C.B., B. McKnight.
DGI: study conception, design, management: L.G.; phenotype collection, data management: P.A.; data analysis: P.A., C. Ladenvall.
FUSION: study conception, design, management: K.L.M., M.B.; phenotype collection, data management: H.M.S., J.T.; data analysis: H.M.S., A.U.J.
Framingham Heart Study: study conception, design, management: R.S.V., E.J.B., D.L.; phenotype collection, data management: S.R.P., R.S.V., S.-J.H., E.J.B., D.L.; data analysis: S.R.P., S.-J.H.
GAIN: study conception, design, management: D.F.L., P.V.G.; phenotype collection, data management: A.R.S., D.F.L., J. Duan, J.S., P.V.G.; sample processing and genotyping: J. Duan, P.V.G.; data analysis: A.R.S., D.F.L., J. Duan, J.S., P.V.G.
IARC/ARCAGE/Central European GWAS: phenotype collection, data management: D.Z., N.S.-D., J.L., P.R., E.F., D.M., V.B., L. Foretova, V.J., S. Benhamou, P.L., I.H., L.R., K.K., A.A., X.C., T.V.M., L. Barzan, C.C., R.L., D.I. Conway, A.Z., C.M.H., P.B.; sample processing and genotyping: J.D.M., M.L., P.B.; data analysis: E.H.L., J.D.M.
InCHIANTI: study conception, design, management: T.M.F., J.M.G., S. Bandinelli; phenotype collection, data management: Y.M.; data analysis: J.R.B.P.
MIGEN: study conception, design, management: R.E., V.S., O.M., C.J.O., D. Altshuler; phenotype collection, data management: G.L., S.M.S., R.E., V.S., B.F.V., O.M., S.K., C.J.O.; sample processing and genotyping: S.K., D. Altshuler; data analysis: G.L., B.F.V., D. Altshuler
NESDA: study conception, design, management: B.W.P., J.H.S.; phenotype collection, data management: B.W.P., J.H.S., N.V.; sample processing and genotyping: B.W.P., J.H.S.; data analysis: N.V.
NTR: study conception, design, management: D.I.B., G.W., E.J.C.d.G.; phenotype collection, data management: D.I.B., G.W., E.J.C.d.G., J.M.V.; sample processing and genotyping: D.I.B., G.W., E.J.C.d.G.; data analysis: J.M.V.
NHS: phenotype collection, data management: S.E.H., D.J.H., P.K., F.G.; sample processing and genotyping: S.J.C., S.E.H., D.J.H., P.K.; data analysis: S.J.C., F.G., P.K.
Rotterdam: study conception, design, management: A.H.; phenotype collection, data management: H.T., A.G.U.; sample processing and genotyping: H.T., A.G.U.; data analysis: H.T., A.G.U., S.W., C.M.v.D.
WGHS: study conception, design, management: B.M.E., G.P., D.I. Chasman, P.M.R.; phenotype collection, data management: B.M.E., G.P., D.I. Chasman, P.M.R.; sample processing and genotyping: G.P., D.I. Chasman; data analysis: B.M.E., G.P., D.I. Chasman.
Corresponding authors
Ethics declarations
Competing interests
The author declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–5, Supplementary Figures 1 and 2 and Supplementary Note (PDF 655 kb)
Supplementary Table 6
Association testing for CPD on chromosome 15, conditional on rs1051730 (XLS 46 kb)
Rights and permissions
About this article
Cite this article
The Tobacco and Genetics Consortium. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet 42, 441–447 (2010). https://doi.org/10.1038/ng.571
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.571
This article is cited by
-
PASTRY: achieving balanced power for detecting risk and protective minor alleles in meta-analysis of association studies with overlapping subjects
BMC Bioinformatics (2024)
-
Unravelling the complex causal effects of substance use behaviours on common diseases
Communications Medicine (2024)
-
The shared genetic architecture of smoking behaviours and psychiatric disorders: evidence from a population-based longitudinal study in England
BMC Genomic Data (2023)
-
A smoking cessation intervention for people with severe mental illness treated in ambulatory mental health care (KISMET): study protocol of a randomised controlled trial
BMC Psychiatry (2023)
-
The association between smoking cessation and lifestyle/genetic variant rs6265 among the adult population in Taiwan
Scientific Reports (2023)