Abstract
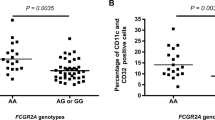
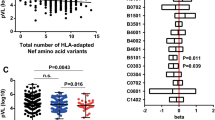
A variant 35 kb upstream of the HLA-C gene (-35C/T) was previously shown to associate with HLA-C mRNA expression level and steady-state plasma HIV RNA levels. We genotyped this variant in 1,698 patients of European ancestry with HIV. Individuals with known seroconversion dates were used for disease progression analysis and those with longitudinal viral load data were used for viral load analysis. We further tested cell surface expression of HLA-C in normal donors using an HLA-C-specific antibody. We show that the -35C allele is a proxy for high HLA-C cell surface expression, and that individuals with high-expressing HLA-C alleles progress more slowly to AIDS and control viremia significantly better than individuals with low HLA-C expressing alleles. These data strongly implicate high HLA-C expression levels in more effective control of HIV-1, potentially through better antigen presentation to cytotoxic T lymphocytes or recognition and killing of infected cells by natural killer cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Carrington, M. & O'Brien, S.J. The influence of HLA genotype on AIDS. Annu. Rev. Med. 54, 535–551 (2003).
Fellay, J. et al. A whole-genome association study of major determinants for host control of HIV-1. Science 317, 944–947 (2007).
Gao, X. et al. Effect of a single amino acid change in MHC class I molecules on the rate of progression to AIDS. N. Engl. J. Med. 344, 1668–1675 (2001).
Gao, X. et al. AIDS restriction HLA allotypes target distinct intervals of HIV-1 pathogenesis. Nat. Med. 11, 1290–1292 (2005).
Flores-Villanueva, P.O. et al. Control of HIV-1 viremia and protection from AIDS are associated with HLA-Bw4 homozygosity. Proc. Natl. Acad. Sci. USA 98, 5140–5145 (2001).
Martin, M.P. et al. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat. Genet. 31, 429–434 (2002).
Martin, M.P. et al. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat. Genet. 39, 733–740 (2007).
Kiepiela, P. et al. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature 432, 769–775 (2004).
Alter, G. et al. Differential natural killer cell-mediated inhibition of HIV-1 replication based on distinct KIR/HLA subtypes. J. Exp. Med. 204, 3027–3036 (2007).
Jin, X. et al. Human immunodeficiency virus type 1 (HIV-1)-specific CD8+-T-cell responses for groups of HIV-1-infected individuals with different HLA-B*35 genotypes. J. Virol. 76, 12603–12610 (2002).
Qi, Y. et al. KIR/HLA pleiotropism: protection against both HIV and opportunistic infections. PLoS Pathog. 2, e79 (2006).
Stranger, B.E. et al. Genome-wide associations of gene expression variation in humans. PLoS Genet. 1, e78 (2005).
Stranger, B.E. et al. Population genomics of human gene expression. Nat. Genet. 39, 1217–1224 (2007).
Braud, V.M., Allan, D.S., Wilson, D. & McMichael, A.J. TAP- and tapasin-dependent HLA-E surface expression correlates with the binding of an MHC class I leader peptide. Curr. Biol. 8, 1–10 (1998).
Shimizu, Y., Geraghty, D.E., Koller, B.H., Orr, H.T. & DeMars, R. Transfer and expression of three cloned human non-HLA-A,B,C class I major histocompatibility complex genes in mutant lymphoblastoid cells. Proc. Natl. Acad. Sci. USA 85, 227–231 (1988).
Apps, R. et al. Human leucocyte antigen (HLA) expression of primary trophoblast cells and placental cell lines, determined using single antigen beads to characterize allotype specificities of anti-HLA antibodies. Immunology 127, 26–39 (2009).
Brodsky, F.M., Bodmer, W.F. & Parham, P. Characterization of a monoclonal anti-beta 2-microglobulin antibody and its use in the genetic and biochemical analysis of major histocompatibility antigens. Eur. J. Immunol. 9, 536–545 (1979).
Barnstable, C.J. et al. Production of monoclonal antibodies to group A erythrocytes, HLA and other human cell surface antigens-new tools for genetic analysis. Cell 14, 9–20 (1978).
Cohen, G.B. et al. The selective downregulation of class I major histocompatibility complex proteins by HIV-1 protects HIV-infected cells from NK cells. Immunity 10, 661–671 (1999).
Rammensee, H., Bachmann, J., Emmerich, N.P., Bachor, O.A. & Stevanovic, S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics 50, 213–219 (1999).
Doytchinova, I.A., Guan, P. & Flower, D.R. Identifiying human MHC supertypes using bioinformatic methods. J. Immunol. 172, 4314–4323 (2004).
Martin, M.P. et al. Genetic acceleration of AIDS progression by a promoter variant of CCR5. Science 282, 1907–1911 (1998).
Smith, M.W. et al. Contrasting genetic influence of CCR2 and CCR5 variants on HIV-1 infection and disease progression. Hemophilia Growth and Development Study (HGDS), Multicenter AIDS Cohort Study (MACS), Multicenter Hemophilia Cohort Study (MHCS), San Francisco City Cohort (SFCC), ALIVE Study. Science 277, 959–965 (1997).
Altfeld, M. et al. Influence of HLA-B57 on clinical presentation and viral control during acute HIV-1 infection. AIDS 17, 2581–2591 (2003).
Fernandez, N.C. et al. A subset of natural killer cells achieves self-tolerance without expressing inhibitory receptors specific for self-MHC molecules. Blood 105, 4416–4423 (2005).
Kim, S. et al. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature 436, 709–713 (2005).
Yawata, M. et al. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J. Exp. Med. 203, 633–645 (2006).
Anfossi, N. et al. Human NK cell education by inhibitory receptors for MHC class I. Immunity 25, 331–342 (2006).
Winter, C.C. & Long, E.O. A single amino acid in the p58 killer cell inhibitory receptor controls the ability of natural killer cells to discriminate between the two groups of HLA-C allotypes. J. Immunol. 158, 4026–4028 (1997).
Phair, J. et al. Acquired immune deficiency syndrome occurring within 5 years of infection with human immunodeficiency virus type-1: the Multicenter AIDS Cohort Study. J. Acquir. Immune Defic. Syndr. 5, 490–496 (1992).
Goedert, J.J. et al. A prospective study of human immunodeficiency virus type 1 infection and the development of AIDS in subjects with hemophilia. N. Engl. J. Med. 321, 1141–1148 (1989).
Buchbinder, S.P., Katz, M.H., Hessol, N.A., O'Malley, P.M. & Holmberg, S.D. Long-term HIV-1 infection without immunologic progression. AIDS 8, 1123–1128 (1994).
Vlahov, D. et al. Prognostic indicators for AIDS and infectious disease death in HIV-infected injection drug users: plasma viral load and CD4+ cell count. J. Am. Med. Assoc. 279, 35–40 (1998).
Emu, B. et al. Phenotypic, functional, and kinetic parameters associated with apparent T-cell control of human immunodeficiency virus replication in individuals with and without antiretroviral treatment. J. Virol. 79, 14169–14178 (2005).
Martin, M.P. & Carrington, M. KIR genotyping and analysis: genotyping and disease association analysis. Methods Mol. Biol. 415, 49–64 (2008).
Council of State and Territorial Epidemiologists. AIDS Program. Center for Infectious Diseases. Revision of the CDC surveillance case definition for acquired immunodeficiency syndrome. MMWR Morb. Mortal. Wkly. Rep. 36 (suppl.1), 1S–15S (1987).
Price, A.L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006).
Apps, R., Gardner, L., Hiby, S.E., Sharkey, A.M. & Moffett, A. Conformation of human leucocyte antigen-C molecules at the surface of human trophoblast cells. Immunology 124, 322–328 (2008).
Menier, C. et al. Characterization of monoclonal antibodies recognizing HLA-G or HLA-E: new tools to analyze the expression of nonclassical HLA class I molecules. Hum. Immunol. 64, 315–326 (2003).
Lee, N., Goodlett, D.R., Ishitani, A., Marquardt, H. & Geraghty, D.E. HLA-E surface expression depends on binding of TAP-dependent peptides derived from certain HLA class I signal sequences. J. Immunol. 160, 4951–4960 (1998).
Tamura, K., Dudley, J., Nei, M. & Kumar, S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599 (2007).
Acknowledgements
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contracts HHSN261200800001E, N02-CP-55504, R01-DA04334 and R01-DA12568. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government. This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. This research was partially funded by a grant from the Bill & Melinda Gates Foundation as part of the Collaboration for AIDS Vaccine Discovery. We would also like to acknowledge the Swiss HIV Cohort Study, supported by the Swiss National Science Foundation, and the SCOPE study was funded by the UL1 RR024131 (Clinical and Translational Sciences Award) and P30 AI27763 (Center for AIDS Research) grants. R.A. is funded by the Cambridge Center for Trophoblast Research. We also thank R. Fernando and the Anthony Nolan Research Institute for the Luminex analysis.
Author information
Authors and Affiliations
Contributions
Author contributions are listed in alphabetical order. Project conception and supervision: M.C.; study rationale: A.M., D.G., D.V.M., J.F., M.C.; data analysis: D.G., Y.Q.; data interpretation and manuscript preparation: M.C., R.A, R.T.; genotyping/sequencing: M.P.M.; R.T.; X.G.; HLA-C expression characterization: G.O.C., R.A., R.T., V.M.; phylogenetic analysis: C.O.H.; clinical samples and data: A.T., B.D.W., G.D.K., J.J.G., J.M., J.N.M., S.B., S.G.D.; manuscript editing: all authors.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1, 2, 3, 4 and 6 and Supplementary Figures 1 and 2 (PDF 359 kb)
Supplementary Table 5
Viral load analysis of 47 SNPs in the HLA-C region. (XLS 68 kb)
Rights and permissions
About this article
Cite this article
Thomas, R., Apps, R., Qi, Y. et al. HLA-C cell surface expression and control of HIV/AIDS correlate with a variant upstream of HLA-C. Nat Genet 41, 1290–1294 (2009). https://doi.org/10.1038/ng.486
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.486
This article is cited by
-
Comparison between qPCR and RNA-seq reveals challenges of quantifying HLA expression
Immunogenetics (2023)
-
IRF7 and RNH1 are modifying factors of HIV-1 reservoirs: a genome-wide association analysis
BMC Medicine (2021)
-
Polymorphism at rs9264942 is associated with HLA-C expression and inflammatory bowel disease in the Japanese
Scientific Reports (2020)
-
Mass spectrometry–based identification of MHC-bound peptides for immunopeptidomics
Nature Protocols (2019)
-
Immunological MHC supertypes and allelic expression: how low is the functional MHC diversity in free-ranging Namibian cheetahs?
Conservation Genetics (2019)