Abstract
Reductive evolution and massive pseudogene formation have shaped the 3.31-Mb genome of Mycobacterium leprae, an unculturable obligate pathogen that causes leprosy in humans. The complete genome sequence of M. leprae strain Br4923 from Brazil was obtained by conventional methods (6× coverage), and Illumina resequencing technology was used to obtain the sequences of strains Thai53 (38× coverage) and NHDP63 (46× coverage) from Thailand and the United States, respectively. Whole-genome comparisons with the previously sequenced TN strain from India revealed that the four strains share 99.995% sequence identity and differ only in 215 polymorphic sites, mainly SNPs, and by 5 pseudogenes. Sixteen interrelated SNP subtypes were defined by genotyping both extant and extinct strains of M. leprae from around the world. The 16 SNP subtypes showed a strong geographical association that reflects the migration patterns of early humans and trade routes, with the Silk Road linking Europe to China having contributed to the spread of leprosy.
Similar content being viewed by others
Main
Leprosy is a chronic dermatological and neurological disease that results from infection with the unculturable pathogen M. leprae1 and causes nerve damage that can lead to severe disabilities. There is no known reservoir for M. leprae other than human beings. New opportunities for understanding the transmission of the leprosy bacillus and its phylogeny have arisen following the determination of the complete 3.3-Mb genome sequence of the TN strain, from Tamil Nadu, India2.
A notable feature of the M. leprae genome is the exceptionally large number of pseudogenes, which occupy almost half of the TN chromosome2. The resulting loss of function most likely accounts for the exceptionally slow growth rate of the bacillus and for researchers' failure to culture it in vitro. Given this extensive genome decay, one might expect to find more genetic variability between different isolates of M. leprae, but initial analysis of SNPs demonstrated that these were very rare, occurring roughly once every 28 kb. Furthermore, all extant isolates of M. leprae were nearly indistinguishable, belonging to one of only four SNP types, and are derived from a single clone3. Variable number tandem repeats (VNTRs) have also been investigated in M. leprae3,4,5,6,7,8 and, in some cases, have proved useful for countrywide epidemiological surveys9. However, owing to variability of VNTR profiles in samples taken from different sites on the same patient, their utility may be limited7,10,11.
The emerging discipline of microbial phylogeography is a powerful means of monitoring not only the spread of microbes but also the movement of their hosts. For instance, compelling associations were found between the genotypes of Helicobacter pylori strains and their places of origin, and the migration and ethnicity of their human hosts12,13,14. M. leprae is also proving useful in this respect, with its spread reflecting the migrations of early humans3, and similar studies with tuberculosis patients suggest that Mycobacterium tuberculosis lineages have also adapted to particular human populations15.
In the present work, the complete genome sequence of Br4923, a Brazilian strain of M. leprae, has been determined and compared with that of the TN isolate, leading to the discovery of a total of only 155 SNPs, of which 78 are informative. To deepen the comparison and avoid possible ascertainment bias16, the genomes of strains from North America and Thailand were resequenced using Illumina (Solexa) technology, revealing comparably low levels of diversity. For phylogeographic purposes, the presence of the 78 informative SNPs was subsequently surveyed in ∼400 isolates, enabling classification of M. leprae into 16 SNP subtypes of limited geographic distribution that correlate with the patterns of human migrations and trade routes.
Results
Complete genome sequence of Br4923
The Br4923 strain of M. leprae was chosen for complete genome analysis because it was originally isolated from a patient in Brazil, the country with the second highest leprosy burden, and because Brazil is geographically remote from India1,17. The genome comprises 3,268,071 bp and is thus 141 bp smaller than that of the TN strain2. No evidence was found for DNA inversions, translocations or duplications, and no transposition or amplification of either the defective insertion sequence elements or the four families of dispersed repeats was detected18, consistent with the findings of an earlier quantitative PCR study3.
On alignment of the two genomes, 194 polymorphic sites were uncovered: these correspond to 155 SNPs, 31 VNTR regions and eight insertion or deletion (indel) events. On verification of the original sequence data, nine SNPs were found to stem from sequencing errors2. The distribution of the true SNPs were as follows: 52 in genes encoding proteins, 39 in pseudogenes, 26 in noncoding regions (or as-yet-unidentified pseudogenes) and 38 in dispersed repeats. The majority of the 31 VNTR regions were found to affect homopolymeric tracts (HPT), di- and trinucleotide repeats, as well as some longer sequences, the longest of which is 52 bp in length and present in two and three copies in the TN and Br4923 strains, respectively. Variation in the di- and trinucleotide repeats is now well documented4,5,8,9,10,11. It is noteworthy that, at the VNTR loci, the repeat copy number was consistently higher in the TN strain than in Br4923, as 23 of the 31 Brazilian VNTR were shorter than their Indian counterparts. Six of the eight indels occurred in repetitive elements and the others were in an intergenic region and the gene ML0825c.
Recombination between dispersed repeats?
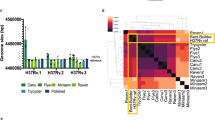
The SNPs associated with dispersed repeats deserve some comment, as they provide evidence for genome plasticity in M. leprae. Variation between different copies of repeat family members had previously been reported18,19, but analysis of two complete genomes provided a richer, more comprehensive dataset. Although all four repeat families (RLEP, REPLEP, LEPRPT and LEPREP) were present in the same copy number and location in both genomes, roughly half of the family members displayed sequence polymorphisms when pairwise comparisons were performed (Fig. 1). The number of polymorphic sites ranged from one in LEPRPT and REPLEP to six in RLEP. With one exception, these resulted from G-A transitions in the RLEP, LEPRPT and LEPREP elements or single-base indels in LEPREP or REPLEP. The polymorphic sites tend to be occupied by A in the TN strain and by G in Br4923. Variation in REPLEP occurs at position 636, which is occupied either by GGG or GG (Fig. 1). Almost 25% of the total SNPs (38/155) occur in these repeats, which account for a mere 1.16% of the genome. The over-representation of SNPs in these elements may indicate that recombination events between different copies of the repetitive elements result in the dispersal of a particular SNP. This interpretation is supported by the strain-specific bias for A and G in the TN and Br4923 strains, respectively, and the finding that more differences are found toward the center of the element rather than near its ends. In turn, these combined findings render polymorphic sites in repetitive DNA unattractive as potential epidemiological tools.
The sites of SNP located within the RLEP, LEPREP, REPLEP and LEPRPT elements present in the genomes of the TN and Br4923 strains are shown and color coded. The position of the SNP is given above and the repeat identifier at left. When occupied by the same nucleotide in both strains, the coloring is continuous: for example, light blue for G. If the same change occurs within a repeat family, a different continuous color is used: for example, light orange for A. When a site is polymorphic between strains, discontinuous darker colors are used. Deletions are identified by *.
Search for informative SNPs
For phylogenetic and phylogeographic purposes, we determined which SNPs had been inherited vertically and which were informative compared to those restricted to a single strain. To do this, seven well-characterized strains of M. leprae (two of which were later resequenced) representing all four known SNP types were examined for the presence of 117 SNPs (that is, all SNPs except those occurring in dispersed repeats). This systematic analysis revealed that 78 SNPs were informative (66%) and 39 SNPs were uninformative (33%).
Identification of informative indels and VNTR
A similar approach was taken to uncover indels and VNTR offering phylogenetic potential, and this resulted in the identification of two informative indels (InDel-1476519 and InDel-978589, TN genome positions) and four informative homopolymeric tracts (HPT-741133, HPT-3244472, HPT-1414666 and HPT-3041556). Given that we had failed to find a reliable relationship between the SNP type and the pattern of six VNTR earlier, and in light of the inherent instability of VNTRs10,11, we did not pursue them further.
Resequencing isolates from North America and Thailand
Owing to the exceptionally high level of genome conservation, M. leprae is particularly suitable for genome resequencing using Illumina technology. Deep coverage was obtained of the genomes of the M. leprae strains Thai53 (38×) and NHDP63 (46×) from Thailand and the United States, respectively, and the reads were mapped onto the TN consensus sequence. The entire genome was covered by the assembly except for the dispersed repeats that could not be distinguished owing to the short read length. This analysis led to the identification of a combined total of 201 SNPs and 14 single-base indels, many of which were shared with the TN or Br4923 strains (Supplementary Table 1), and it uncovered five pseudogenes (Fig. 2a). A distance matrix was then established (Fig. 2b) and used to construct a phylogenetic tree by neighbor joining20. The topology of the tree (Fig. 2c) is fully consistent with the previous scheme derived by SNP typing3.
(a) Polymorphisms associated with new pseudogenes with the position referring to the TN genome. (b) Diversity matrix based on polymorphisms found in nonrepetitive sequences. Note that there are 21, 23, 65 and 56 SNP and/or single-base indels restricted to the TN, Thai53, NHDP63 and Br4923 strains, respectively. (c) Phylogenetic tree from neighbor joining based on polymorphisms from b.
Synonymous and nonsynonymous substitutions
Determining the ratio of nonsynonymous to synonymous SNPs (dN/dS) is a popular method used to predict whether any purifying selection is operating21. When the four genomes were compared, we found 43 (42%) synonymous and 59 (58%) nonsynonymous changes in the protein-coding genes (Supplementary Table 1); these are values similar to those reported for the M. tuberculosis complex (38% and 62%, respectively), in which SNPs are >10-fold more abundant22. The dN/dS ratios were calculated within the M. leprae genes through pairwise comparisons revealing an average value of 0.70. A ratio close to 1 is indicative of no, or weak, selection against nonsynonymous SNPs as recently described for Salmonella typhi (with a dN/dS = 0.66)23. Like S. typhi, another obligate human pathogen, M. leprae also seems to show little genetic drift, probably because of its small effective population size21. Phylogenetic trees were constructed using the nonsynonymous and synonymous SNPs24, and these had similar topology to that shown in Figure 2c.
Phylogeographic survey of extant M. leprae
In our previous study, we had genotyped 175 strains of M. leprae from 21 different geographic origins but found only four phylogenetic groups: SNP-types 1–4 (ref. 3). A close relationship was established between the geographical origin of the strain and its genotype, giving a first impression of how leprosy may have disseminated around the globe as humans migrated. However, certain parts of the world were unrepresented, particularly the Middle East and Europe. Because these regions were of historical importance in the migration of human populations—notably the Middle East region, where early humans are thought to have arrived from Africa and from whence the European and Asian migrations began25—we made intensive efforts to obtain samples from these settings. Samples were obtained from Iran and Turkey in the Middle East and from China, Korea and Japan in the Far East.
The 84 informative polymorphic sites were used in our survey (Fig. 3a, Supplementary Table 2), and these sites enabled us to reconstruct the movement of leprosy between peoples and countries. A total of 400 samples from 28 regions were successfully screened, resulting in the definition of 16 different M. leprae subtypes, as compared to the four originally described3; these are referred to as A–P (where A corresponds to the TN isolate and Br4923 is P, Fig. 3a). SNP type 1 is now subdivided into four subtypes (A–D), SNP type 2 into four subtypes (E–H), SNP type 3 into five subtypes (I–M) and SNP type 4 into three subtypes (N–P). For instance, subtypes A and B differ from each other at one HPT and 21 SNP. Likewise, although there are no SNP differences within members of SNP type 4, these can be subdivided based on differences in their indel or HPT profiles.
(a) The panel shows the four SNP types (1–4) and the 16 subtypes (A–P) used to classify strains of M. leprae. Classification is based on SNP (green), HPT (blue) and indels (orange), with the bases indicated representing one of only two possibilities found. Indels are denoted by −. The bases shown are those associated with a given genotype. For instance, subtypes 1A and 1B differ by 21 SNP and 1 HPT, for a total difference in 22 biodiversity markers. Note that for each subtype the markers are present en bloc. (b) Scheme explaining how the ancestral versions of polymorphic sites were deduced by comparison with an outgroup, in this case M. tuberculosis. (c) Phylogenetic tree made by neighbor joining based on extant M. leprae sequences and rooted with the ancestral sequence from b and Supplementary Table 2.
To exclude the possible effects of ascertainment bias16, we challenged this evolutionary model by phylogenetic analysis using other actinobacteria, such as M. tuberculosis, as an outgroup. Inspection of the positions corresponding to the M. leprae SNPs in other genome sequences allowed the likely ancestral base to be deduced (Fig. 3b, Supplementary Table 2). This was successful for 13 of the 15 groups of markers and enabled us to establish a consensus ancestral sequence, which was then used in either neighbor-joining or maximum likelihood analysis to produce a phylogenetic tree (Fig. 3c). The topology of the trees was fully consistent with that of the scheme and phylogenetic tree for the four fully sequenced M. leprae strains (Figs. 2c and 3a), thereby confirming its robustness.
An additional phylogenetic analysis was performed using maximum likelihood analysis26,27 to probe the relationship between the genotype of the M. leprae isolates and the affected individual's country of origin. For completeness, an example of each genotype found within a country was included, resulting in a sample size of 61. Overall, there was a consistent trend in the relationship between genotype and the geographic region (Fig. 4), and this trend gives an indication of the likely direction of gene flow and dissemination of the leprosy bacillus.
The tree was rooted between genotypes 2 and 3, and a single member of each genotype present per country was included. For each replicate, tree leaves were attributed to 1 of 11 broad geographical locations and a maximum likelihood inference of ancestral (geographical) states is shown. Branch lengths are arbitrary, and nodes with bootstrap support values above 60 are indicated together with the distribution of ancestral states at each internal node. Vertical bars indicate major geographical locations for each cluster.
The global distribution of the 16 SNP subtypes was then plotted to show the frequency of each genotype per country. Again, with the exception of islands3, this plot showed a reasonably tight correlation between thecountry of origin of an individual, the M. leprae genotype of the strain infecting the individual, and the known pattern of human migrations (Fig. 5). It is of interest that the M. leprae strains from both Turkey and Iran fall into the same two groups, F and K. This is consistent with F being a precursor to strains that migrated eastward with human populations, later giving rise to SNP type A found in India and Southeast Asia, whereas group K may have been an ancestor for M. leprae associated with westward migrations of humans that led to genotype M, associated with Europe and the Americas, and genotype P, characteristic of South America. Findings with Chinese isolates of M. leprae9 are also important, as, unlike many samples from Southeast Asia, these are not genotype 1 but genotype 3, subtype K.
Pillars are located on the country of origin of the M. leprae sample and color coded according to the scheme for the 16 SNP subtypes shown in Figure 3 (note that this differs from the color scheme in Fig. 4). The thickness of the pillar corresponds to the number of samples (1–5, thin; 6–29, intermediate; >30, broad). The gray arrows indicate the migration routes of humans, with the estimated time of migration in years shown25,46. The red dots indicate the location of the Silk Road in the first century, and * denotes results obtained from a-DNA.
Phylogeography of ancient M. leprae
Studying ancient DNA (a-DNA) is a valuable yet challenging approach, as this not only enables us to obtain samples from countries where leprosy is extinct but also provides information about different time periods and past epidemics. Skeletal remains were obtained from leprosy graveyards in Croatia, Denmark, Egypt, England, Hungary and Turkey, and all showed clear osteological evidence of lepromatous leprosy (Table 1). These remains were first shown to be positive for M. leprae DNA by single-round RLEP PCR28 and then were used to generate PCR products for sequence analysis of all three SNP typing loci and, when possible, the appropriate subtypes. In all 13 cases, the a-DNA samples were found to be of SNP type 3, and 7 of these were successfully subtyped. The a-DNA analysis revealed that M. leprae from the UK belonged to SNP subtype 3I, whereas samples from Hungary were of SNP subtypes 3K or 3M (Table 1). Both extinct and extant samples of M. leprae from Turkey belonged to type 3K, whereas the 1,500-year-old Egyptian sample was found to be SNP type 3. All of the archaeological cases exhibited different VNTR profiles, which assisted in authentication of the a-DNA analysis (data not shown).
Discussion
Here, we describe the complete genome sequence and comparative analysis of Br4923, a Brazilian strain of M. leprae, and its use for the discovery of SNPs and other polymorphic markers with phylogeographic potential. This finding was then complemented by genome resequencing of strains from Thailand and the United States. When the four genome sequences were compared, remarkably little genomic diversity was uncovered, consistent with the hypothesis that leprosy has arisen from infection with a single clone that has passed through a recent evolutionary bottleneck2,3. The four strains, which came from widely separated countries, have genomes that are 99.995% identical. In terms of the diagnosis, treatment and prevention of leprosy, this is extremely encouraging, as it means that antigenic drift in M. leprae should be negligible and the sequences of drug targets will not vary. Indeed, only 49 of the estimated 1,614 proteins in M. leprae show any amino acid change (Supplementary Table 1).
In a recent comparative study with M. tuberculosis and Streptomyces coelicolor, it was estimated that most of the mutations (56%) associated with pseudogenes or noncoding regions in M. leprae could be attributed to transitions, mainly of the C→T or G→A types29. When the noncoding or pseudogene-containing regions were compared in the two completely sequenced M. leprae genomes, the frequency of these transitional mutations (59%) was found to be very close to that value, whereas in the coding sequences, the frequency was 63.5%. As outlined above, the same mutations are also dominant in the repetitive elements, where they account for 97% of the changes. However, as these elements appear to undergo recombination (Fig. 1), this particular value may be misleading. Nonetheless, a decrease in the G+C content of a genome is thought to be a hallmark of reductive evolution30,31, and at 57.8% M. leprae clearly conforms to this rule, as the G+C content of most sequenced mycobacterial genomes is ∼66%.
Five new pseudogenes were uncovered by this work (Fig. 2a), and their orthologs in M. tuberculosis are all known or predicted to be nonessential32. The most noteworthy of these pseudogenes was found in Br4923, where the counterpart of ML0825c in the TN strain has acquired a frameshift mutation as the result of a thymidine insertion in codon 16 (TN position 978629). ML0825c encodes a transcriptional regulator belonging to the ArsR family that is predicted to repress target gene expression in the absence of heavy metal cations. In M. tuberculosis, the ML0825c ortholog rv2358 is the proximal gene in a bicistronic operon with zur (furB), whose expression is repressed by rv2358 in a zinc-dependent manner33,34. In an M. tuberculosis zur mutant, expression of 32 genes was found to be upregulated35. It is not known whether loss of ML0825c has any phenotypic consequences, but it is conceivable that, as a result of polarity from the frameshift mutation, Zur may not be made, thus leading to derepression of the Zur regulon. This mutation is found only in M. leprae strains belonging to SNP subtypes 4O and 4P. The present study has also uncovered another useful marker for SNP subtype 4P, Del-1476519, a single-base deletion restricted to M. leprae strains from South America or the Caribbean. We can conclude that loss of the corresponding thymidine must have occurred within the last 500 years, since the introduction to Brazil of the ancestral strain from West Africa during the slave trade.
Through genome comparisons, some authors have attempted to reconstruct the evolution of M. leprae and to predict whether pseudogene formation was a gradual or stochastic event36,37. Using a new approach based on the number of nonsynonymous substitutions per site in pseudogenes, one group estimated that M. leprae and M. tuberculosis diverged 66 million years ago and that a single pseudogenization event must have occurred in the leprosy bacillus in the last 10–20 million years36. Although we cannot predict the precise date of pseudogene formation here, our data indicate that this process still occurs.
The main goal of this study was to discover new polymorphic markers in order to improve the resolution of epidemiological and phylogeographic studies of M. leprae. This was successfully achieved, and interrogation of the 84 informative sites has led to the 4 SNP types3 being resolved into 16 subtypes. Together with an increase in the number of countries surveyed, this study has resulted in a much deeper level of understanding of the global spread of leprosy, with confirmation of the relationship between SNP type and geographical origin of the strains.
Paleomicrobiology was particularly helpful in this respect, and the analysis of ancient M. leprae DNA, present in skeletal remains from countries where leprosy has been eradicated, not only enabled us to expand our geographical coverage but also provided insight into the genotypes of strains circulating in Europe, Turkey and Egypt as long as 1,500 years ago. From the initial scheme for the evolution of the different M. leprae genotypes, it was predicted that European isolates should belong to SNP type 3, and this was indeed found to be the case (Table 1). Thanks to the new tools developed here, it was also possible to subtype the isolates in some cases, despite the limiting amounts and extensive fragmentation of the a-DNA. Examination of B116 (Table 1), the Egyptian skeleton from the Kellis-2 site in the Dakhleh Oasis38, deserves some comment, as this specimen comes from a region close to the proposed origin of both M. leprae and Homo sapiens. Based on genotype data from East African strains of M. leprae, one might expect to find SNP type 2 in Egypt, whereas in reality we found type 3. B116, who has a calibrated radiocarbon date of 445 ± 50 years AD, in the Roman period, was buried in close proximity to another individual with leprosy, termed B6. Dietary studies using 15N stable isotopic analysis showed that both burials were outliers compared to other inhumations at Kellis 2 and may therefore not have been of Egyptian origin. Alternatively, from the phylogeny shown in Figure 3c, it is possible that when the ancestor of M. leprae separated from the other mycobacteria, its genotype was between SNP types 2 and 3.
The present phylogeographic scheme, based on results obtained with ∼400 samples of M. leprae from 28 different locations, extends considerably the hypothesis proposed earlier in which the progenitor strain may have originated in East Africa and was SNP type 2 (ref. 3). This then gave rise to SNP type 1, which spread eastward with humans into Asia and SNP type 3, which disseminated westward into the Middle East and Europe before spawning the type 4 strains that are found in West Africa and countries linked to West Africa by the slave trade. Two new conclusions can be drawn from interpretation of the refined scheme (Figs. 3,4,5).
First, leprosy appears to have been introduced into Asia by two different routes: a southern route, associated with the SNP type 1 strains encountered in the Indian subcontinent, Indonesia and the Philippines, and a more northerly route starting in the Eastern Mediterranean region and extending via Turkey and Iran to China and from there to Korea and Japan. Here, strains with the 3K SNP subtype are preponderant, and the trade route between Europe and Asia known as the Silk Road appears likely to have been a means of transport and disease transmission (Fig. 5). The Black Death, caused by Yersinia pestis39, is thought to have reached Europe in the fourteenth century from China via the Silk Road, carried by humans and their fleas. For leprosy (Fig. 3a) the opposite route of transmission may have operated, with the disease originating in Europe or the Middle East and then spreading to the Far East. Leprosy was thought to have reached China from India, in about 500 BC, and then to have spread from China to Japan40. This proposition is incompatible with the data presented here.
Second, it seems unlikely that leprosy was introduced into the Americas by early humans via the Bering straits; rather, it appears more probable that it was brought by immigrants from Europe, as most of the M. leprae strains found in North, Central and South America have the 3I genotype found in European leprosy cases. This interpretation is consistent with paleological findings because skeletons with signs of leprosy are limited to the postcolonial period41.
Finally, it is worth discussing the enormous discrepancy between the period at which pseudogene formation is thought to have arisen and the origin of early humans. It has been estimated recently that the bulk of the pseudogenes in M. leprae arose no earlier than 9 million years ago36. Pseudogene formation is an indicator of radical change in the lifestyle of the host bacterium, such as from the free-living to pathogenic state or of adaptation to life within a particular tissue or cell type30,31. In the case of M. leprae, obligate parasitism of humans or another primate species would represent such a change. Although modern humans represented by H. sapiens have existed only since approximately 250,000 years ago and left Africa within the last 100,000 years to settle other regions, earlier hominids are thought to have diverged from chimpanzees over 5 million years ago42. Reconciliation of the estimated time of pseudogene formation with human evolution could be achieved if an ancestor of M. leprae infected an early primate and then underwent genome decay and was subsequently transmitted vertically—although this seems unlikely, given that more genetic diversity among M. leprae isolates would be expected if this were true. Alternatively, the genome decay could well be ancient, but M. leprae may only recently have become a human pathogen. For instance, it is conceivable that an ancestral form of M. leprae infected an invertebrate host such as an insect, which later acted as a vector for transmitting the bacillus to humans. Support for the latter scenario is provided by studies of the related pathogen Mycobacterium ulcerans, which is at an early stage of reductive evolution43 and appears to be transmitted to humans by water bugs and/or mosquitoes44,45. Further insight into the timing of pseudogene formation in M. leprae will be provided by microbiology and paleomicrobiology and by deeper genome sequence analysis.
Methods
Complete sequence of Br4923 and informatics.
Br4923 was originally isolated in 1996 from the skin biopsy of a Brazilian patient, inoculated into nude mice, and its whole genome sequence obtained from a pcDNA2.1 shotgun library. Briefly, 5 μg of purified DNA was sheared by nebulization and cloned in the pcDNA2.1 vector using adapters as described previously50. The sequencing template was obtained with a Templiphi kit (GE Healthcare), subjected to BigDye Terminator version 3.1 cycle sequencing method and run in an ABI3730 DNA sequencer (Applied Biosystems). The sequence was assembled from 21,300 reads, analyzed and annotated using the Staden package (Trev, Gap4), Blast, Act and Artemis51,52,53,54,55, as described previously2,56.
Genome resequencing of Thai53 and NHDP63 and informatics.
The strains Thai53 and NHDP63 were originally from an individual from Thailand and a native-born American from Louisiana, United States, respectively. Genomic DNA fragment sequencing libraries were prepared using the DNA Sample Prep Kit (Illumina) according to the protocol supplied with the reagents and using 5 μg of genomic DNA. DNA fragment libraries were loaded into one (Thai53) or two lanes (NHDP63) of a flow cell and sequenced on the Genome Analyzer II (Illumina) using the 36 Cycle Sequencing Kit version 1. Data were processed using the Illumina Pipeline Software package version 1.0 and reads either mapped onto the TN consensus sequence using MAQ57 or assembled using Edena58 and aligned to the reference genome using blastn55. Data were managed using GAP4 and variations reported by direct comparisons.
Phylogeny.
Multiple alignments were generated using ClustalW59 and phylogenetic trees calculated by the neighbor-joining or maximum likelihood methods using web-based software (see URL). To calculate dN/dS ratios, we used the START 2 package60 with the Nei-Gojobori method and the Jukes-Cantor correction61. For the phylogeographic analysis, we selected one SNP per linkage disequilibrium block (n = 25) for each combination of geographical location and genotype (n = 61), excluding samples from islands. These sequences were input to the RaxML software26,27, which produced a maximum likelihood tree and 200 bootstrap replicates. Trees were rooted between genotypes 2 and 3. For each replicate, the tree leaves were attributed 1 of 11 broad geographical locations and a maximum likelihood inference of ancestral (geographical) states was performed using the ape package under R62. The inferred transition rates were averaged over all replicates and applied to the maximum likelihood tree (Fig. 4).
Sample preparation from extant M. leprae.
Details of the specimens examined and their origin may be found in Supplementary Table 3. In this study, we performed PCR amplification with >600 samples of M. leprae comprising purified DNA, fresh skin biopsies, paraffin-embedded biopsy samples and slit-skin smears from microscope slides. M. leprae cells and/or DNA were prepared differently according to the sample source. Fresh skin biopsies were treated as described previously and DNA released by 'freeze boiling'63. Paraffin-embedded specimens were first heated at 60 °C for 3 h, then the paraffin was removed by two extractions with 1 ml of xylene for 15 min, which was followed by one extraction with 100% ethanol for 15 min. Tissue was progressively rehydrated by soaking in 30% ethanol and, finally, in water. Then biopsies were minced with fine scissors followed by shearing in a Qiagen lyser with 3 mm glass beads (10 min, maximum power). The supernatant was transferred to an Eppendorf tube and 'freeze boiled'. After removal of debris, the supernatant was used directly in PCR. For slit-skin samples we used the Qiaamp DNA micro kit (Qiagen, Inc.) to recover DNA from stained microscope slides.
Standard PCR, SNP analysis and sequence reactions.
The seven well-characterized strains of M. leprae used as sources of DNA for initial work were TN, India 2 and Thai53 (all SNP type 1), Africa (SNP type 2), NHDP63 and NHDP98 (both SNP type 3) and Br4923 (SNP type 4)3. Details of the primers used for genotyping may be found in Supplementary Table 4. Reactions (20 μl) typically contained M. leprae DNA from different samples, 10 mM Tris-HCL (pH 9.0), 50 mM KCl, 1.5 mM MgCl2, 1.25 U Taq DNA polymerase (Q-Biogene) and 200 nM of forward and reverse primers. PCR was carried out for 45 cycles consisting of denaturation at 94 °C for 1 min, annealing at 55 °C for 1 min and extension at 72 °C for 2 min, with a final extension at 72 °C for 10 min in a thermocycler (PTC-100, MJ Research, Inc.). If amplification failed, the PCR reaction was repeated after addition of 1 μl of T4GP32 (Q-Biogene). After enzymatic treatment with Exonuclease I (USB, Corp.) and Shrimp Alkaline Phosphatase (USB, Corp.), PCR products were submitted to BigDye Terminator version 3.1 cycle sequencing and analyzed using an ABI3100 DNA sequencer (Applied Biosystems). Sequence data were analyzed as above51,54.
Ancient DNA (a-DNA) studies.
Details of samples taken for the bioarchaeological part of the study are shown in Table 1. Measures to prevent crossover contamination26 were followed from the time of sampling. The strategy followed included (i) the use of multiple extraction and template blanks, (ii) reproducibility, (iii) appropriate molecular behavior, (iv) confirmation of product identity with sequencing and (v) replication of key findings at a separate center. One of the samples, burial 1914 from Ipswich, was sampled twice to provide sufficient material for analysis at center 2, Manchester University. Gloves were worn and changed between handling different skeletal components. Samples of bone were removed from the skeleton using sterile disposable scalpel blades and transferred into sterile plastic containers for transport to the laboratories. The work surface was cleaned between samples using a proprietary multisurface cleaner containing bleach.
a-DNA extraction, PCR and sequencing.
At center 1, University College London, samples of bone or scrapings were finely ground in autoclaved pestles and mortars and DNA from bone powder extracted using the NucliSens kit from bioMérieux, as previously described28. DNA extracts were stored at –20 °C until assayed. At center 2, Manchester University, DNA from burial 1914 was extracted independently using a modification of the method of Yang64, which has previously been shown to be the most efficient of five tested methods for extraction of a-DNA from bones65.
The presence of residual M. leprae DNA was first confirmed using a sensitive PCR method, which amplifies the 37-copy repetitive element RLEP18,19,28. SNP typing methods were developed to amplify fragmented DNA likely to persist in skeletal remains and applied to the extracts. The oligonucleotide primers used for this and the key PCR conditions are listed in Supplementary Table 5.
The Excite core kit (BioGene) was used for all PCR amplifications. This is a uracil-N-glycosylase-ready kit suitable for real-time and routine hot-start PCR applications. SYBR green was included in the PCR master mixes at a final dilution of 1/55,000, and reactions were performed and monitored on the Corbett RotorGene 3000 real-time PCR platform (Corbett Research) in a final volume of 25 μl. Forty-five cycles of amplification were performed for all methods. Melt analysis was performed using the RotorGene software and, additionally, all products were run on 3% agarose gels. Template blanks containing water in place of bone extract were alternated with samples in the RotorGene chamber and monitored to ensure absence of contamination. Positive control samples were not amplified during the course of the bioarchaeological study.
PCR products were separated on 3% (wt/vol) low-melting-point agarose (Invitrogen), and bands were excised with a sterile scalpel blade and purified using a Geneclean III DNA isolation kit (Fisher Life Sciences), then sequenced using BigDye Terminators, as outlined above.
URLs.
Mobyle@Pasteur, http://mobyle.pasteur.fr; NCBI short read archive, http://www.ncbi.nlm.nih.gov/Traces/sra_sub/sub.cgi.
Accession codes.
Br4923 genome sequence, GenBank FM211192; reads from Thai53 and NHDP63 resequencing analysis, SRP001064 (trace depository).
Note: Supplementary information is available on the Nature Genetics website.
Accession codes
Change history
05 February 2010
In the version of this article initially published 1 November 2009, Ali Khamesipour was misspelled in the author list. The error has been corrected in the HTML and PDF versions of the article.
References
Britton, W.J. & Lockwood, D.N. Leprosy. Lancet 363, 1209–1219 (2004).
Cole, S.T. et al. Massive gene decay in the leprosy bacillus. Nature 409, 1007–1011 (2001).
Monot, M. et al. On the origin of leprosy. Science 308, 1040–1042 (2005).
Groathouse, N.A. et al. Multiple polymorphic loci for molecular typing of strains of Mycobacterium leprae. J. Clin. Microbiol. 42, 1666–1672 (2004).
Shin, Y.C. et al. Variable numbers of TTC repeats in Mycobacterium leprae DNA from leprosy patients and use in strain differentiation. J. Clin. Microbiol. 38, 4535–4538 (2000).
Truman, R., Fontes, A.B., De Miranda, A.B., Suffys, P. & Gillis, T. Genotypic variation and stability of four variable-number tandem repeats and their suitability for discriminating strains of Mycobacterium leprae. J. Clin. Microbiol. 42, 2558–2565 (2004).
Young, S.K. et al. Microsatellite mapping of Mycobacterium leprae populations in infected humans. J. Clin. Microbiol. 42, 4931–4936 (2004).
Zhang, L., Budiawan, T. & Matsuoka, M. Diversity of potential short tandem repeats in Mycobacterium leprae and application for molecular typing. J. Clin. Microbiol. 43, 5221–5229 (2005).
Weng, X. et al. Identification and distribution of Mycobacterium leprae genotypes in a region of high leprosy prevalence in China: a 3-year molecular epidemiological study. J. Clin. Microbiol. 45, 1728–1734 (2007).
Monot, M. et al. Are variable-number tandem repeats appropriate for genotyping Mycobacterium leprae? J. Clin. Microbiol. 46, 2291–2297 (2008).
Young, S.K. et al. Use of short tandem repeat sequences to study Mycobacterium leprae in leprosy patients in Malawi and India. PLoS Negl. Trop. Dis. 2, e214 (2008).
Falush, D. et al. Traces of human migrations in Helicobacter pylori populations. Science 299, 1582–1585 (2003).
Linz, B. et al. An African origin for the intimate association between humans and Helicobacter pylori. Nature 445, 915–918 (2007).
Wirth, T. et al. Distinguishing human ethnic groups by means of sequences from Helicobacter pylori: lessons from Ladakh. Proc. Natl. Acad. Sci. USA 101, 4746–4751 (2004).
Gagneux, S. et al. Variable host-pathogen compatibility in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 103, 2869–2873 (2006).
Achtman, M. Evolution, population structure, and phylogeography of genetically monomorphic bacterial pathogens. Annu. Rev. Microbiol. 62, 53–70 (2008).
World Health Organization. Global leprosy situation, 2007. Wkly. Epidemiol. Rec. 82, 225–232 (2007).
Cole, S.T., Supply, P. & Honoré, N. Repetitive sequences in Mycobacterium leprae and their impact on genome plasticity. Lepr. Rev. 72, 449–461 (2001).
Woods, S.A. & Cole, S.T. A family of dispersed repeats in Mycobacterium leprae. Mol. Microbiol. 4, 1745–1751 (1990).
Saitou, N. & Nei, M. The neighbour-joining method: a new method for constructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 (1987).
Rocha, E.P. et al. Comparisons of dN/dS are time dependent for closely related bacterial genomes. J. Theor. Biol. 239, 226–235 (2006).
Hershberg, R. et al. High functional diversity in Mycobacterium tuberculosis driven by genetic drift and human demography. PLoS Biol. 6, e311 (2008).
Holt, K.E. et al. High-throughput sequencing provides insights into genome variation and evolution in Salmonella typhi. Nat. Genet. 40, 987–993 (2008).
Baker, L., Brown, T., Maiden, M.C. & Drobniewski, F. Silent nucleotide polymorphisms and a phylogeny for Mycobacterium tuberculosis. Emerg. Infect. Dis. 10, 1568–1577 (2004).
Underhill, P.A. et al. Y chromosome sequence variation and the history of human populations. Nat. Genet. 26, 358–361 (2000).
Stamatakis, A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 (2006).
Stamatakis, A., Hoover, P. & Rougemont, J. A rapid bootstrap algorithm for the RAxML Web servers. Syst. Biol. 57, 758–771 (2008).
Taylor, G.M., Watson, C.L., Lockwood, D.N.J. & Mays, S.A. Variable nucleotide tandem repeat (VNTR) typing of two cases of lepromatous leprosy from the archaeological record. J. Archaeol. Sci. 33, 1569–1579 (2006).
Mitchell, A. & Graur, D. Inferring the pattern of spontaneous mutation from the pattern of substitution in unitary pseudogenes of Mycobacterium leprae and a comparison of mutation patterns among distantly related organisms. J. Mol. Evol. 61, 795–803 (2005).
Parkhill, J. et al. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat. Genet. 35, 32–40 (2003).
Parkhill, J. et al. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413, 523–527 (2001).
Sassetti, C.M., Boyd, D.H. & Rubin, E.J. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48, 77–84 (2003).
Milano, A., Branzoni, M., Canneva, F., Profumo, A. & Riccardi, G. The Mycobacterium tuberculosis Rv2358-furB operon is induced by zinc. Res. Microbiol. 155, 192–200 (2004).
Canneva, F., Branzoni, M., Riccardi, G., Provvedi, R. & Milano, A. Rv2358 and FurB: two transcriptional regulators from Mycobacterium tuberculosis which respond to zinc. J. Bacteriol. 187, 5837–5840 (2005).
Maciag, A. et al. Global analysis of the Mycobacterium tuberculosis Zur (FurB) regulon. J. Bacteriol. 189, 730–740 (2007).
Gómez-Valero, L., Rocha, E.P., Latorre, A. & Silva, F.J. Reconstructing the ancestor of Mycobacterium leprae: the dynamics of gene loss and genome reduction. Genome Res. 17, 1178–1185 (2007).
Madan Babu, M. Did the loss of sigma factors initiate pseudogene accumulation in M. leprae? Trends Microbiol. 11, 59–61 (2003).
Molto, J.E. in The Past and Present of Leprosy: Archaeological, Historical, Palaeopathological and Clinical Approaches, BAR International Series 1054 (eds. Roberts, C.A., Lewis, M.E. & Manchester, K.) 179–192 (Archaeopress, Oxford, UK, 2002).
Achtman, M. et al. Microevolution and history of the plague bacillus, Yersinia pestis. Proc. Natl. Acad. Sci. USA 101, 17837–17842 (2004).
Browne, S.G. in Leprosy. (ed. Hastings, R.C.) 1–14 (Churchill Livingstone, Edinburgh, UK, 1985).
Ortner, D.J. in The Identification of Pathological Conditions in Human Skeletal Remains (ed. Ortner, D.J.) 227–272 (Academic Press, London, UK, 2003).
Strait, D.S., Grine, F.E. & Moniz, M.A. A reappraisal of early hominid phylogeny. J. Hum. Evol. 32, 17–82 (1997).
Stinear, T.P. et al. Reductive evolution and niche adaptation inferred from the genome of Mycobacterium ulcerans, the causative agent of Buruli ulcer. Genome Res. 17, 192–200 (2007).
Johnson, P.D. et al. Mycobacterium ulcerans in mosquitoes captured during outbreak of Buruli ulcer, southeastern Australia. Emerg. Infect. Dis. 13, 1653–1660 (2007).
Marsollier, L. et al. Aquatic insects as a vector for Mycobacterium ulcerans. Appl. Environ. Microbiol. 68, 4623–4628 (2002).
Cavalli-Sforza, L.L. & Feldman, M.W. The application of molecular genetic approaches to the study of human evolution. Nat. Genet. 33 (Suppl.), 266–275 (2003).
Pálfi, G. et al. in The Past and Present of Leprosy: Archaeological, Historical, Palaeopathological and Clinical Approaches. BAR International Series 1054 (eds. Roberts, C.A., Lewis, M.E. & Manchester, K.) 205–209 (Archaeopress, Oxford, UK, 2002).
Molnar, E., Marcsik, A., Bereczki, Z. & Donoghue, H.D. Pathological cases from the 7th century in Hungary 16th European Meeting of the Paleopathology Association, Paleopathology Newsletter Supplement. (Fira Santorini, Greece, 2006).
Erdal, Y.S. Health status of the Kovuklukaya (Boyabat/Sinop) people and its relations with their lifestyle. Anadolu Arastirmalari (in Turkish) 22, 169–196 (2004).
Garnier, T. et al. The complete genome sequence of Mycobacterium bovis. Proc. Natl. Acad. Sci. USA 100, 7877–7882 (2003).
Bonfield, J.K., Smith, K.F. & Staden, R. A new DNA sequence assembly program. Nucleic Acids Res. 24, 4992–4999 (1995).
Carver, T.J. et al. ACT: the Artemis Comparison Tool. Bioinformatics 21, 3422–3423 (2005).
Rutherford, K. et al. Artemis: sequence visualization and annotation. Bioinformatics 16, 944–945 (2000).
Altschul, S.F., Gish, W., Miller, W., Myers, E.W. & Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Altschul, S.F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997).
Stinear, T.P. et al. Insights from the complete genome sequence of Mycobacterium marinum on the evolution of Mycobacterium tuberculosis. Genome Res. 18, 729–741 (2008).
Li, H., Ruan, J. & Durbin, R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18, 1851–1858 (2008).
Hernandez, D., Francois, P., Farinelli, L., Osteras, M. & Schrenzel, J. De novo bacterial genome sequencing: millions of very short reads assembled on a desktop computer. Genome Res. 18, 802–809 (2008).
Thompson, J.D., Higgins, D.G. & Gibson, T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994).
Jolley, K.A., Feil, E.J., Chan, M.S. & Maiden, M.C. Sequence type analysis and recombinational tests (START). Bioinformatics 17, 1230–1231 (2001).
Nei, M. & Gojobori, T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3, 418–426 (1986).
Paradis, E., Claude, J. & Strimmer, K. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics 20, 289–290 (2004).
Woods, S.A. & Cole, S.T. A rapid method for the detection of potentially viable Mycobacterium leprae in human biopsies: a novel application of PCR. FEMS Microbiol. Lett. 53, 305–309 (1989).
Yang, D.Y., Waye, J.S., Dudar, J.C. & Saunders, S.R. Technical note: improved DNA extraction from ancient bones using silica-based spin columns. Am. J. Phys. Anthropol. 105, 539–543 (1998).
Bouwman, A.S. & Brown, T.A. Comparison between silica-based methods for the extraction of DNA from human bones from 18th–19th century London. Anc. Biomol. 4, 173–178 (2002).
Acknowledgements
We thank the study participants and staff from the various leprosy clinics for their invaluable participation and support; J. Molto, A. Marcsik, Y. Selim Erdal, E. Popescu, M. Slaus and J. Boldsen for their encouragement and help with a-DNA samples; and L. Frangeul, A. Kapopoulou and S. Uplekar for assistance with bioinformatics. This work received the financial support of the Fondation Raoul Follereau, the Génopole programme, and the US National Institutes of Health, National Institute of Allergy and Infectious Diseases (grant RO1-AI47197-01A1 and contract NO1-AI25469).
Author information
Authors and Affiliations
Contributions
M.M., N.H., P.J.B. and S.T.C. designed the study; A.P.-M, M. Matsuoka., A.K., Y.D., S.J., T.H.R., L.V.-C., M.M.S, S.B., M. Macdonald, B.R.S. and J.S.S. contributed sources of M. leprae DNA and demographic information; N.H., M. Monot, N.Z., D.S., T.G., J.T., K.H, A.G., J.R., P.S. and P.B. performed DNA sequencing and bioinformatics; G.M.T., H.D.D., A.B., S.M., C.W. and D.L. provided, sequenced and analyzed a-DNA and contributed archaeological insight; and S.T.C. wrote the manuscript with contributions from M. Monot., N.H. and G.M.T. plus comments from all authors.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–5 (PDF 367 kb)
Rights and permissions
About this article
Cite this article
Monot, M., Honoré, N., Garnier, T. et al. Comparative genomic and phylogeographic analysis of Mycobacterium leprae. Nat Genet 41, 1282–1289 (2009). https://doi.org/10.1038/ng.477
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.477
This article is cited by
-
Leprosy in wild chimpanzees
Nature (2021)
-
Real-time PCR-based quantitation of viable Mycobacterium leprae strain from clinical samples and environmental sources and its genotype in multi-case leprosy families of India
European Journal of Clinical Microbiology & Infectious Diseases (2020)
-
Leprosy Transmission in Amazonian Countries: Current Status and Future Trends
Current Tropical Medicine Reports (2020)
-
The recombination-cold region as an epidemiological marker of recombinogenic opportunistic pathogen Mycobacterium avium
BMC Genomics (2019)
-
DNA gyrase could be a crucial regulatory factor for growth and survival of Mycobacterium leprae
Scientific Reports (2019)