Abstract
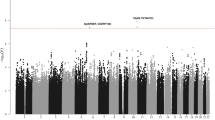
Using the Affymetrix 500K Mapping array and publicly available genotypes, we identified 18 SNPs whose allele frequency differed significantly(P < 1 × 10−5) between pediatric acute lymphoblastic leukemia (ALL) cases (n = 317) and non-ALL controls (n = 17,958). Two SNPs in ARID5B not only differed between ALL and non-ALL groups (rs10821936, P = 1.4 × 10−15, odds ratio (OR) = 1.91; rs10994982, P = 5.7 × 10−9, OR = 1.62) but also distinguished B-hyperdiploid ALL from other subtypes (rs10821936, P = 1.62 × 10−5, OR = 2.17; rs10994982, P = 0.003, OR 1.72). These ARID5B SNPs also distinguished B-hyperdiploid ALL from other subtypes in an independent validation cohort (n = 124 children with ALL; P = 0.003 and P = 0.0008, OR 2.45 and 2.86, respectively) and were associated with methotrexate accumulation and gene expression pattern in leukemic lymphoblasts. We conclude that germline variants affect susceptibility to, and characteristics of, specific ALL subtypes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
Accession codes
References
Pui, C.H. & Evans, W.E. Treatment of acute lymphoblastic leukemia. N. Engl. J. Med. 354, 166–178 (2006).
Schnakenberg, E. et al. Polymorphisms of methylenetetrahydrofolate reductase (MTHFR) and susceptibility to pediatric acute lymphoblastic leukemia in a German study population. BMC Med. Genet. 6, 23 (2005).
Urayama, K.Y. et al. MDR1 gene variants, indoor insecticide exposure, and the risk of childhood acute lymphoblastic leukemia. Cancer Epidemiol. Biomarkers Prev. 16, 1172–1177 (2007).
Wiemels, J.L. et al. Methylenetetrahydrofolate reductase (MTHFR) polymorphisms and risk of molecularly defined subtypes of childhood acute leukemia. Proc. Natl. Acad. Sci. USA 98, 4004–4009 (2001).
Stanulla, M. et al. Polymorphisms within glutathione S-transferase genes (GSTM1, GSTT1, GSTP1) and risk of relapse in childhood B-cell precursor acute lymphoblastic leukemia: a case-control study. Blood 95, 1222–1228 (2000).
Healy, J. et al. Promoter SNPs in G1/S checkpoint regulators and their impact on the susceptibility to childhood leukemia. Blood 109, 683–692 (2007).
Mathonnet, G., Krajinovic, M., Labuda, D. & Sinnett, D. Role of DNA mismatch repair genetic polymorphisms in the risk of childhood acute lymphoblastic leukaemia. Br. J. Haematol. 123, 45–48 (2003).
Paulsson, K. et al. Evidence for a single-step mechanism in the origin of hyperdiploid childhood acute lymphoblastic leukemia. Genes Chromosom. Cancer 44, 113–122 (2005).
Kager, L. et al. Folate pathway gene expression differs in subtypes of acute lymphoblastic leukemia and influences methotrexate pharmacodynamics. J. Clin. Invest. 115, 110–117 (2005).
Nordgard, S.H. et al. Genome-wide analysis identifies 16q deletion associated with survival, molecular subtypes, mRNA expression, and germline haplotypes in breast cancer patients. Genes Chromosom. Cancer 47, 680–696 (2008).
Sobti, R.C. et al. Combined effect of GSTM1, GSTT1 and GSTP1 polymorphisms on histological subtypes of lung cancer. Biomarkers 13, 282–295 (2008).
Kim, M.S. et al. Frameshift mutation of UVRAG, an autophagy-related gene, in gastric carcinomas with microsatellite instability. Hum. Pathol. 39, 1059–1063 (2008).
Campbell, P.J. Somatic and germline genetics at the JAK2 locus. Nat. Genet. 41, 385–386 (2009).
Greaves, M. Childhood leukaemia. Br. Med. J. 324, 283–287 (2002).
Chen, H.C. et al. Genetic polymorphisms of metabolic enzymes CYP1A1, CYP2D6, GSTM1 and GSTT1 and leukemia susceptibility. Eur. J. Cancer Prev. 17, 251–258 (2008).
Pui, C.H. & Evans, W.E. Acute lymphoblastic leukemia. N. Engl. J. Med. 339, 605–615 (1998).
Pui, C.H., Relling, M.V. & Downing, J.R. Acute lymphoblastic leukemia. N. Engl. J. Med. 350, 1535–1548 (2004).
Ziino, O. et al. Acute lymphoblastic leukemia in children with associated genetic conditions other than Down's syndrome. The AIEOP experience. Haematologica 91, 139–140 (2006).
Greaves, M.F. Speculations on the cause of childhood acute lymphoblastic leukemia. Leukemia 2, 120–125 (1988).
Perera, F.P. Environment and cancer: who are susceptible? Science 278, 1068–1073 (1997).
Dong, L.M. et al. Genetic susceptibility to cancer: the role of polymorphisms in candidate genes. J. Am. Med. Assoc. 299, 2423–2436 (2008).
Patsialou, A., Wilsker, D. & Moran, E. DNA-binding properties of ARID family proteins. Nucleic Acids Res. 33, 66–80 (2005).
Lahoud, M.H. et al. Gene targeting of Desrt, a novel ARID class DNA-binding protein, causes growth retardation and abnormal development of reproductive organs. Genome Res. 11, 1327–1334 (2001).
Wilsker, D., Patsialou, A., Dallas, P.B. & Moran, E. ARID proteins: a diverse family of DNA binding proteins implicated in the control of cell growth, differentiation, and development. Cell Growth Differ. 13, 95–106 (2002).
Wilsker, D. et al. The DNA-binding properties of the ARID-containing subunits of yeast and mammalian SWI/SNF complexes. Nucleic Acids Res. 32, 1345–1353 (2004).
Bourquin, J.P. et al. Identification of distinct molecular phenotypes in acute megakaryoblastic leukemia by gene expression profiling. Proc. Natl. Acad. Sci. USA 103, 3339–3344 (2006).
Chang, L.W. et al. Computational identification of the normal and perturbed genetic networks involved in myeloid differentiation and acute promyelocytic leukemia. Genome Biol. 9, R38 (2008).
Molnár, A. & Georgopoulos, K. The Ikaros gene encodes a family of functionally diverse zinc finger DNA-binding proteins. Mol. Cell. Biol. 14, 8292–8303 (1994).
Molnár, A. et al. The Ikaros gene encodes a family of lymphocyte-restricted zinc finger DNA binding proteins, highly conserved in human and mouse. J. Immunol. 156, 585–592 (1996).
Mullighan, C.G. et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature 446, 758–764 (2007).
Mullighan, C.G. et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature 453, 110–114 (2008).
Den Boer, M.L. et al. A subtype of childhood acute lymphoblastic leukaemia with poor treatment outcome: a genome-wide classification study. Lancet Oncol. 10, 125–134 (2009).
Mullighan, C.G. et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N. Engl. J. Med. 360, 470–480 (2009).
Pui, C.H. et al. Improved outcome for children with acute lymphoblastic leukemia: results of Total Therapy Study XIIIB at St Jude Children's Research Hospital. Blood. 104, 2690–2696 (2004).
Pui, C.-H. Treating childhood acute lymphoblastic leukemia without cranial irradiation. N. Engl. J. Med. 360, 2730–2741 (2009).
Borowitz, M.J. et al. Clinical significance of minimal residual disease in childhood acute lymphoblastic leukemia and its relationship to other prognostic factors: a Children's Oncology Group study. Blood 111, 5477–5485 (2008).
The Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447, 661–678 (2007).
Dick, D.M. et al. Genomewide linkage analyses of bipolar disorder: a new sample of 250 pedigrees from the National Institute of Mental Health Genetics Initiative. Am. J. Hum. Genet. 73, 107–114 (2003).
McInnis, M.G. et al. Genome-wide scan and conditional analysis in bipolar disorder: evidence for genomic interaction in the National Institute of Mental Health genetics initiative bipolar pedigrees. Biol. Psychiatry 54, 1265–1273 (2003).
Suarez, B.K. et al. Genomewide linkage scan of 409 European-ancestry and African American families with schizophrenia: suggestive evidence of linkage at 8p23.3-p21.2 and 11p13.1-q14.1 in the combined sample. Am. J. Hum. Genet. 78, 315–333 (2006).
Borowitz, M.J. et al. Minimal residual disease detection in childhood precursor-B-cell acute lymphoblastic leukemia: relation to other risk factors. A Children's Oncology Group study. Leukemia 17, 1566–1572 (2003).
Fabre, G. et al. In vitro formation of polyglutamyl derivatives of methotrexate and 7- hydroxymethotrexate in human lymphoblastic leukemia cells. Cancer Res. 43, 4648–4652 (1983).
Masson, E. et al. Accumulation of methotrexate polyglutamates in lymphoblasts is a determinant of antileukemic effects in vivo. A rationale for high- dose methotrexate. J. Clin. Invest. 97, 73–80 (1996).
Synold, T.W. et al. Blast cell methotrexate-polyglutamate accumulation in vivo differs by lineage, ploidy, and methotrexate dose in acute lymphoblastic leukemia. J. Clin. Invest. 94, 1996–2001 (1994).
Ross, M.E. et al. Classification of pediatric acute lymphoblastic leukemia by gene expression profiling. Blood 102, 2951–2959 (2003).
Yeoh, E.J. et al. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell 1, 133–143 (2002).
Pritchard, J.K., Stephens, M. & Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 155, 945–959 (2000).
Acknowledgements
We thank our protocol co-investigators, clinical and research staff (particularly P. McGill, J. Cai, S.-H. Chen, N. Kornegay and J. Pauley), the Hartwell Center for Bioinformatics and Biotechnology at St. Jude Children's Research Hospital (Memphis, Tennessee), S. Das at the Genetic Services Laboratory at the University of Chicago (Chicago, Illinois) and the study participants and their families who participated. We thank R. Whitson from City of Hope Hospital (Duarte, California) for constructive discussions.
This study makes use of data generated by the Wellcome Trust Case Control Consortium (WTCCC). A full list of the investigators who contributed to the generation of the data is available from http://www.wtccc.org.uk. Funding for the project was provided by the Wellcome Trust under award 076113.
Funding support for the Genome-Wide Association of Schizophrenia Study was provided by the National Institute of Mental Health (R01 MH67257, R01 MH59588, R01 MH59571, R01 MH59565, R01 MH59587, R01 MH60870, R01 MH59566, R01 MH59586, R01 MH61675, R01 MH60879, R01 MH81800, U01 MH46276, U01 MH46289 U01 MH46318, U01 MH79469, and U01 MH79470) and the genotyping of samples was provided through the Genetic Association Information Network (GAIN). The datasets used for the analyses described in this manuscript were obtained from the database of Genotype and Phenotype database (dbGaP) found at http://www.ncbi.nlm.nih.gov/gap through dbGaP (accession number phs000021.v1.p1). Samples and associated phenotype data for the Genome-Wide Association of Schizophrenia Study were provided by the Molecular Genetics of Schizophrenia Collaboration (Principal Investigator: P.V. Gejman, Evanston Northwestern Healthcare (ENH) and Northwestern University, Evanston, Illinois).
Funding support for the Whole Genome Association Study of Bipolar Disorder was provided by the US National Institute of Mental Health (NIMH) and the genotyping of samples was provided through the Genetic Association Information Network (GAIN). The datasets used for the analyses described in this manuscript were obtained from dbGaP (accession number phs000017.v1.p1). Samples and associated phenotype data for the Collaborative Genomic Study of Bipolar Disorder were provided by The NIMH Genetics Initiative for Bipolar Disorder. Data and biomaterials were collected in four projects that participated in the NIMH Bipolar Disorder Genetics Initiative. From 1991 to 1998, the principal investigators and co-investigators were: Indiana University, Indianapolis, Indiana, U01 MH46282, J. Nurnberger, M. Miller and E. Bowman; Washington University, St. Louis, Missouri, U01 MH46280, T. Reich, A. Goate and J. Rice; Johns Hopkins University, Baltimore, Maryland, U01 MH46274, J.R. DePaulo Jr., S. Simpson and C. Stine; NIMH Intramural Research Program, Clinical Neurogenetics Branch, Bethesda, Maryland, E. Gershon, D. Kazuba and E. Maxwell. Data and biomaterials were collected as part of ten projects that participated in the NIMH Bipolar Disorder Genetics Initiative. From 1999 to 2003, the principal investigators and co-investigators were: Indiana University, Indianapolis, Indiana, R01 MH59545, J. Nurnberger, M.J. Miller, E.S. Bowman, N.L. Rau, P.R. Moe, N. Samavedy, R. El-Mallakh (University of Louisville, Louisville, Kentucky), H. Manji (Wayne State University, Detroit, Michigan), D.A. Glitz (Wayne State University), E.T. Meyer, C. Smiley, T. Foroud, L. Flury, D.M. Dick and H. Edenberg; Washington University, St. Louis, Missouri, R01 MH059534, J. Rice, T. Reich, A. Goate and L. Bierut; Johns Hopkins University, Baltimore, Maryland, R01 MH59533, M. McInnis, J.R. DePaulo Jr., D.F. MacKinnon, F.M. Mondimore, J.B. Potash, P.P. Zandi, D. Avramopoulos and J. Payne; University of Pennsylvania, Philadelphia, Pennsylvania, R01 MH59553, W. Berrettini; University of California, Irvine, California, R01 MH60068, W. Byerley and M. Vawter; University of Iowa, Iowa City, Iowa, R01 MH059548, W. Coryell and R. Crowe; University of Chicago, Chicago, Illinois, R01 MH59535, E. Gershon, J. Badner, F. McMahon, C. Liu, A. Sanders, M. Caserta, S. Dinwiddie, T. Nguyen and D. Harakal; University of California, San Diego, California, R01 MH59567, J. Kelsoe and R. McKinney; Rush University, Chicago, Illinois, R01 MH059556, W. Scheftner, H.M. Kravitz, D. Marta, A. Vaughn-Brown and L. Bederow; NIMH Intramural Research Program, Bethesda, Maryland, 1Z01MH002810-01, F.J. McMahon, L. Kassem, S. Detera-Wadleigh, L. Austin and D.L. Murphy.
This study was supported by the US National Cancer Institute (grants CA 51001, CA 078224, CA 36401 and CA 21765), the National Institute of Health/National Institutes of General Medical Sciences Pharmacogenetics Research Network and Database (U01 GM61393, U01 HL65899, U01GM61374; http://www.pharmgkb.org/), by a Center of Excellence grant from the State of Tennessee and by the American Lebanese Syrian Associated Charities (ALSAC).
Author information
Authors and Affiliations
Contributions
L.R.T., W.Y. and M.V.R designed the study. M.V.R. and W.E.E. directed the study. L.R.T., W.Y., D.F., S.P.H., W.L.C., M.D., C.W., G.N., J.D., S.C.R., C.-H.P, W.E.E. and M.V.R. helped with data analysis and data interpretation. W.Y. conducted the statistical genomic analysis. S.R. performed and guided cytogenetic data analysis. S.H., W.L.C., M.D. and C.-H.P. provided clinical data. J.D. provided leukemic lymphoblast gene expression data.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–10, Supplementary Figures 1–5 and a Supplementary Note (PDF 979 kb)
Rights and permissions
About this article
Cite this article
Treviño, L., Yang, W., French, D. et al. Germline genomic variants associated with childhood acute lymphoblastic leukemia. Nat Genet 41, 1001–1005 (2009). https://doi.org/10.1038/ng.432
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.432
This article is cited by
-
Genome-wide trans-ethnic meta-analysis identifies novel susceptibility loci for childhood acute lymphoblastic leukemia
Leukemia (2022)
-
Enrichment of cancer-predisposing germline variants in adult and pediatric patients with acute lymphoblastic leukemia
Scientific Reports (2022)
-
Lymphoblastic Lymphoma: a Concise Review
Current Oncology Reports (2022)
-
Relationship between IKZF1 polymorphisms and the risk of acute lymphoblastic leukemia: a meta-analysis*
Oncology and Translational Medicine (2022)
-
The CEBPE rs2239633 genetic polymorphism on susceptibility to childhood acute lymphoblastic leukemia: an updated meta-analysis
Environmental Health and Preventive Medicine (2021)