Abstract
Asthma, hay fever (or allergic rhinitis) and eczema (or atopic dermatitis) often coexist in the same individuals1, partly because of a shared genetic origin2,3,4. To identify shared risk variants, we performed a genome-wide association study (GWAS; n = 360,838) of a broad allergic disease phenotype that considers the presence of any one of these three diseases. We identified 136 independent risk variants (P < 3 × 10−8), including 73 not previously reported, which implicate 132 nearby genes in allergic disease pathophysiology. Disease-specific effects were detected for only six variants, confirming that most represent shared risk factors. Tissue-specific heritability and biological process enrichment analyses suggest that shared risk variants influence lymphocyte-mediated immunity. Six target genes provide an opportunity for drug repositioning, while for 36 genes CpG methylation was found to influence transcription independently of genetic effects. Asthma, hay fever and eczema partly coexist because they share many genetic risk variants that dysregulate the expression of immune-related genes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Pinart, M. et al. Comorbidity of eczema, rhinitis, and asthma in IgE-sensitised and non-IgE-sensitised children in MeDALL: a population-based cohort study. Lancet Respir. Med. 2, 131–140 (2014).
Thomsen, S.F. et al. Findings on the atopic triad from a Danish twin registry. Int. J. Tuberc. Lung Dis. 10, 1268–1272 (2006).
van Beijsterveldt, C.E. & Boomsma, D.I. Genetics of parentally reported asthma, eczema and rhinitis in 5-yr-old twins. Eur. Respir. J. 29, 516–521 (2007).
Loh, P.R. et al. Contrasting genetic architectures of schizophrenia and other complex diseases using fast variance-components analysis. Nat. Genet. 47, 1385–1392 (2015).
Yang, J. et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet. 44, 369–375, S1–S3 (2012).
Ferreira, M.A. Improving the power to detect risk variants for allergic disease by defining case–control status based on both asthma and hay fever. Twin Res. Hum. Genet. 17, 505–511 (2014).
Bulik-Sullivan, B.K. et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015).
GTEx Consortium. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–660 (2015).
Wells, A. et al. The anatomical distribution of genetic associations. Nucleic Acids Res. 43, 10804–10820 (2015).
Finucane, H.K. et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 47, 1228–1235 (2015).
Farh, K.K. et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 518, 337–343 (2015).
Fehrmann, R.S. et al. Gene expression analysis identifies global gene dosage sensitivity in cancer. Nat. Genet. 47, 115–125 (2015).
Thomsen, S.F., Kyvik, K.O. & Backer, V. Etiological relationships in atopy: a review of twin studies. Twin Res. Hum. Genet. 11, 112–120 (2008).
Sanseau, P. et al. Use of genome-wide association studies for drug repositioning. Nat. Biotechnol. 30, 317–320 (2012).
Bonder, M.J. et al. Disease variants alter transcription factor levels and methylation of their binding sites. Nat. Genet. 49, 131–138 (2017).
Joehanes, R. et al. Epigenetic signatures of cigarette smoking. Circ Cardiovasc Genet 9, 436–447 (2016).
Lev, S. et al. Identification of a novel family of targets of PYK2 related to Drosophila retinal degeneration B (rdgB) protein. Mol. Cell. Biol. 19, 2278–2288 (1999).
Yan, S.R. & Novak, M.J. β2 integrin–dependent phosphorylation of protein-tyrosine kinase Pyk2 stimulated by tumor necrosis factor α and fMLP in human neutrophils adherent to fibrinogen. FEBS Lett. 451, 33–38 (1999).
Kamen, L.A., Schlessinger, J. & Lowell, C.A. Pyk2 is required for neutrophil degranulation and host defense responses to bacterial infection. J. Immunol. 186, 1656–1665 (2011).
Willer, C.J., Li, Y. & Abecasis, G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010).
Fadista, J., Manning, A.K., Florez, J.C. & Groop, L. The (in)famous GWAS P-value threshold revisited and updated for low-frequency variants. Eur. J. Hum. Genet. 24, 1202–1205 (2016).
Gough, H. et al. Allergic multimorbidity of asthma, rhinitis and eczema over 20 years in the German birth cohort MAS. Pediatr. Allergy Immunol. 26, 431–437 (2015).
Mortz, C.G., Andersen, K.E., Dellgren, C., Barington, T. & Bindslev-Jensen, C. Atopic dermatitis from adolescence to adulthood in the TOACS cohort: prevalence, persistence and comorbidities. Allergy 70, 836–845 (2015).
Sarnowski, C. et al. Identification of a new locus at 16q12 associated with time to asthma onset. J. Allergy Clin. Immunol. 138, 1071–1080 (2016).
Dharmage, S.C. et al. Atopic dermatitis and the atopic march revisited. Allergy 69, 17–27 (2014).
Chang, X. & Wang, K. wANNOVAR: annotating genetic variants for personal genomes via the web. J. Med. Genet. 49, 433–436 (2012).
1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 (2012).
Rebhan, M., Chalifa-Caspi, V., Prilusky, J. & Lancet, D. GeneCards: integrating information about genes, proteins and diseases. Trends Genet. 13, 163 (1997).
Davis, J.R. et al. An efficient multiple-testing adjustment for eQTL studies that accounts for linkage disequilibrium between variants. Am. J. Hum. Genet. 98, 216–224 (2016).
Montgomery, S.B. et al. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature 464, 773–777 (2010).
Lappalainen, T. et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature 501, 506–511 (2013).
Westra, H.J. et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet. 45, 1238–1243 (2013).
Chun, S. et al. Limited statistical evidence for shared genetic effects of eQTLs and autoimmune-disease-associated loci in three major immune-cell types. Nat. Genet. 49, 600–605 (2017).
Sanyal, A., Lajoie, B.R., Jain, G. & Dekker, J. The long-range interaction landscape of gene promoters. Nature 489, 109–113 (2012).
Mifsud, B. et al. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat. Genet. 47, 598–606 (2015).
Li, G. et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell 148, 84–98 (2012).
Rao, S.S. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014).
Corradin, O. et al. Combinatorial effects of multiple enhancer variants in linkage disequilibrium dictate levels of gene expression to confer susceptibility to common traits. Genome Res. 24, 1–13 (2014).
Hnisz, D. et al. Super-enhancers in the control of cell identity and disease. Cell 155, 934–947 (2013).
He, B., Chen, C., Teng, L. & Tan, K. Global view of enhancer–promoter interactome in human cells. Proc. Natl. Acad. Sci. USA 111, E2191–E2199 (2014).
Andersson, R. et al. An atlas of active enhancers across human cell types and tissues. Nature 507, 455–461 (2014).
Welter, D. et al. The NHGRI GWAS Catalog, a curated resource of SNP–trait associations. Nucleic Acids Res. 42, D1001–D1006 (2014).
Astle, W.J. et al. The allelic landscape of human blood cell trait variation and links to common complex disease. Cell 167, 1415–1429.e19 (2016).
Zheng, J. et al. LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics 33, 272–279 (2017).
Fairfax, B.P. et al. Genetics of gene expression in primary immune cells identifies cell type–specific master regulators and roles of HLA alleles. Nat. Genet. 44, 502–510 (2012).
Fairfax, B.P. et al. Innate immune activity conditions the effect of regulatory variants upon monocyte gene expression. Science 343, 1246949 (2014).
Fu, J. et al. Unraveling the regulatory mechanisms underlying tissue-dependent genetic variation of gene expression. PLoS Genet. 8, e1002431 (2012).
Zhernakova, D.V. et al. Identification of context-dependent expression quantitative trait loci in whole blood. Nat. Genet. 49, 139–145 (2017).
Willemsen, G. et al. The Netherlands Twin Register biobank: a resource for genetic epidemiological studies. Twin Res. Hum. Genet. 13, 231–245 (2010).
Tsai, P.C. et al. DNA methylation changes in the IGF1R gene in birth weight discordant adult monozygotic twins. Twin Res. Hum. Genet. 18, 635–646 (2015).
Acknowledgements
This research was conducted using the UK Biobank resource under application number 10074. Detailed acknowledgments and funding details are provided for each contributing study in the Supplementary Note.
Author information
Authors and Affiliations
Consortia
Contributions
Data collection and analysis in the contributing studies: M.A.F., M.C.M., S.C.D., L.M.B., P.J.T., N.G.M., D.L.D. (AAGC study); J.M.V., G.H.K. (LifeLines study); H.B., E.R., M.H., A.F., N.N., H.S., S.K., C.G., K.S., S.W. (GENEVA study); I.M., F.R., J.E.-G., S.G., A.A., G.H., C.O.S., N.H., Y.-A.L. (GENUFAD studies); C.T., D.A.H. (23andMe study); J.D.H., J.S.W., R.B.M., E.J. (GERA study); Q.H., J.-J.H., G.W., D.I.B. (NTR study); A.T., V.U., Y.L., P.K.E.M., C.A., R.K. (CATSS, TWINGENE and SALTY studies); L.P. (ALSPAC study); B.M.B., L.G.F., M.E.G., J.B.N., W.Z., K.H., A.L., O.L.H., M.L., G.R.A., C.J.W. (HUNT study); L.P., M.A.F. (UK Biobank study). Methylation analysis: J.v.D., D.I.B., R.J. Biological and drug annotation: M.A.F., C.W.M., E.M., K.B., O.H., J.Z., J.A.R., J.B., B.B. Quality control, meta-analysis, tables and figures: M.A.F. Writing group: M.A.F., J.M.V., I.M., C.T., J.D.H., Q.H., A.T., V.U., J.v.D., Y.L., J.E.-G., B.M.B., J.B., S.C.D., S.W., P.K.E.M., R.J., E.J., Y.-A.L., D.I.B., C.A., R.K., G.H.K., L.P. Study design and management: M.A.F., D.A.H., B.M.B., S.W., P.K.E.M., R.J., E.J., Y.-A.L., D.I.B., C.A., R.K., G.H.K., L.P.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
A full list of members and affiliations appears in the Supplementary Note.
A full list of members and affiliations appears in the Supplementary Note.
A full list of members and affiliations appears in the Supplementary Note.
A full list of members and affiliations appears in the Supplementary Note.
Integrated supplementary information
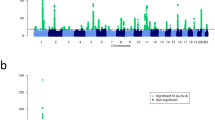
Supplementary Figure 1 Summary of main analyses and key findings.
Meta-analysis of GWAS results from 13 studies identified 136 variants independently associated with disease risk at a P < 3 × 10−8, of which 73 were in low LD (r2 < 0.05) with published allergy risk variants. Based on a high LD (r2 > 0.8) between the 136 sentinel risk variants and sentinel cis-eQTLs and/or nonsynonymous coding variants, a total of 132 likely target genes were identified. The likely target genes were preferentially expressed in whole blood and lung, and enriched among pathways related to lymphocyte immunity. Twenty-nine genes are targets of drugs considered for clinical development, including six for which the effect on gene expression of the allergy-protective allele and the respective drug matched. Thirty-six genes have a nearby CpG for which methylation levels are associated with gene expression levels independently of SNP effects on expression and methylation. For one of these genes, variation in methylation levels at the expression-associated CpG was significantly associated with smoking status.
Supplementary Figure 2 Distribution of the observed and expected association P values for the allergic disease GWAS performed in each individual study that contributed to the meta-analysis.
For each study, the genomic inflation factor (λ; estimated as the median χ2 divided by 0.4549) is also shown. The intercept of LD score regression for each study is shown in Supplementary Table 1.
Supplementary Figure 3 Eighty-nine allergy risk variants (gray bars) in low LD (r2 < 0.05) with each other reported in previous GWAS, according to the year each association was first reported.
We identified 185 SNP associations with allergic disease at P < 5 × 10−8 in the NHGRI-EBI GWAS catalog. Correlated SNPs were then grouped based on LD, such that the lead SNP in each group was in LD (r2 > 0.05) with other variants in that group but not with the lead SNPs of all other groups. This procedure is described in greater detail in the Supplementary Note. This resulted in 89 groups of SNPs associated with allergic disease. The earliest year an association was reported with a variant in each group was identified and plotted. The red bar shows the number of novel variants discovered in this study (50 in new loci and 23 in known loci).
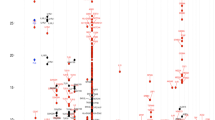
Supplementary Figure 4 Twenty-six of the 136 sentinel variants were significantly associated with variation in the reported age of onset for allergic disease.
We first tested the association between each sentinel variant and the age at which symptoms of any allergic disease (asthma, hay fever or eczema) were first reported, using data from the UK Biobank study (n = 35,972). After correcting for multiple testing, 26 variants (yellow circles) had a significant association with age of onset with the allergy-predisposing allele always associated with decreased age of onset (i.e., a negative β, shown on the y axis). An additional 47 variants (blue circles) were nominally associated (P < 0.05) with age of onset. We then performed the same analysis separately for individuals who reported suffering only from a single disease and formally compared the SNP effects between the three groups. In these analyses, the effect on age of onset was significantly different (P < 0.05) between the three diseases for 8 of the 26 variants (yellow circles with black inner dot), consistent with the presence of disease-specific SNP effects on age of onset.
Supplementary Figure 5 Enrichment of tissue-specific gene expression in 25 broad tissues studied by the GTEx Consortium, after restricting the background gene list to the subset of genes with eQTLs.
We repeated the tissue-specific enrichment analysis described in Figure 3a after restricting the background gene list to the subset of 12,804 genes with a known eQTL. Random genes were also drawn from the subset with a known eQTL.
Supplementary Figure 6 Enrichment of SNP heritability among immune cell enhancers remains after excluding from the analysis the 136 sentinel variants.
We repeated the SNP heritability enrichment analysis described in Figure 3b after excluding the 136 sentinel variants (and all variants correlated at r2 > 0.05) from the meta-analysis results.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 and Supplementary Note
Supplementary Tables
Supplementary Tables 1–29
Rights and permissions
About this article
Cite this article
Ferreira, M., Vonk, J., Baurecht, H. et al. Shared genetic origin of asthma, hay fever and eczema elucidates allergic disease biology. Nat Genet 49, 1752–1757 (2017). https://doi.org/10.1038/ng.3985
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3985
This article is cited by
-
Association between interleukin-6-174G/C gene polymorphism and asthma severity: exploring the role of total serum IgE, blood eosinophils, and FeNO as markers of type 2 inflammation
Allergy, Asthma & Clinical Immunology (2024)
-
A systematic two-sample and bidirectional MR process highlights a unidirectional genetic causal effect of allergic diseases on COVID-19 infection/severity
Journal of Translational Medicine (2024)
-
Genetics of chronic respiratory disease
Nature Reviews Genetics (2024)
-
SCGB1D2 inhibits growth of Borrelia burgdorferi and affects susceptibility to Lyme disease
Nature Communications (2024)
-
A genome-wide association study provides insights into the genetic etiology of 57 essential and non-essential trace elements in humans
Communications Biology (2024)