Abstract
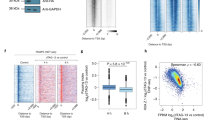
RNA polymerase II (Pol II) pauses downstream of the transcription initiation site before beginning productive elongation. This pause is a key component of metazoan gene expression regulation. Some promoters have a strong disposition for Pol II pausing and often mediate faster, more synchronous changes in expression. This requires multiple rounds of transcription and thus cannot rely solely on pause release. However, it is unclear how pausing affects the initiation of new transcripts during consecutive rounds of transcription. Using our recently developed ChIP-nexus method, we find that Pol II pausing inhibits new initiation. We propose that paused Pol II helps prevent new initiation between transcription bursts, which may reduce noise.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Sainsbury, S., Bernecky, C. & Cramer, P. Structural basis of transcription initiation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 16, 129–143 (2015).
Darzacq, X. et al. Imaging transcription in living cells. Annu. Rev. Biophys. 38, 173–196 (2009).
Nogales, E., Louder, R.K. & He, Y. Cryo-EM in the study of challenging systems: the human transcription pre-initiation complex. Curr. Opin. Struct. Biol. 40, 120–127 (2016).
Conaway, R.C. & Conaway, J.W. General initiation factors for RNA polymerase II. Annu. Rev. Biochem. 62, 161–190 (1993).
Roeder, R.G. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci. 21, 327–335 (1996).
Tirode, F., Busso, D., Coin, F. & Egly, J.M. Reconstitution of the transcription factor TFIIH: assignment of functions for the three enzymatic subunits, XPB, XPD, and cdk7. Mol. Cell 3, 87–95 (1999).
Holstege, F.C., van der Vliet, P.C. & Timmers, H.T. Opening of an RNA polymerase II promoter occurs in two distinct steps and requires the basal transcription factors IIE and IIH. EMBO J. 15, 1666–1677 (1996).
Adelman, K. & Lis, J.T. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat. Rev. Genet. 13, 720–731 (2012).
Jonkers, I. & Lis, J.T. Getting up to speed with transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 16, 167–177 (2015).
Hendrix, D.A., Hong, J.W., Zeitlinger, J., Rokhsar, D.S. & Levine, M.S. Promoter elements associated with RNA Pol II stalling in the Drosophila embryo. Proc. Natl. Acad. Sci. USA 105, 7762–7767 (2008).
Nechaev, S. et al. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science 327, 335–338 (2010).
Lagha, M. et al. Paused Pol II coordinates tissue morphogenesis in the Drosophila embryo. Cell 153, 976–987 (2013).
Boettiger, A.N. & Levine, M. Synchronous and stochastic patterns of gene activation in the Drosophila embryo. Science 325, 471–473 (2009).
Gilchrist, D.A. et al. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell 143, 540–551 (2010).
Zeitlinger, J. et al. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat. Genet. 39, 1512–1516 (2007).
Li, J. et al. Kinetic competition between elongation rate and binding of NELF controls promoter-proximal pausing. Mol. Cell 50, 711–722 (2013).
Core, L.J., Waterfall, J.J. & Lis, J.T. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322, 1845–1848 (2008).
Kwak, H., Fuda, N.J., Core, L.J. & Lis, J.T. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science 339, 950–953 (2013).
Rhee, H.S. & Pugh, B.F. Comprehensive genome-wide protein–DNA interactions detected at single-nucleotide resolution. Cell 147, 1408–1419 (2011).
Pugh, B.F. & Venters, B.J. Genomic organization of human transcription initiation complexes. PLoS One 11, e0149339 (2016).
He, Q., Johnston, J. & Zeitlinger, J. ChIP-nexus enables improved detection of in vivo transcription factor binding footprints. Nat. Biotechnol. 33, 395–401 (2015).
Titov, D.V. et al. XPB, a subunit of TFIIH, is a target of the natural product triptolide. Nat. Chem. Biol. 7, 182–188 (2011).
Geiger, J.H., Hahn, S., Lee, S. & Sigler, P.B. Crystal structure of the yeast TFIIA/TBP/DNA complex. Science 272, 830–836 (1996).
Tan, S., Hunziker, Y., Sargent, D.F. & Richmond, T.J. Crystal structure of a yeast TFIIA/TBP/DNA complex. Nature 381, 127–151 (1996).
Imbalzano, A.N., Zaret, K.S. & Kingston, R.E. Transcription factor (TF) IIB and TFIIA can independently increase the affinity of the TATA-binding protein for DNA. J. Biol. Chem. 269, 8280–8286 (1994).
He, Y. et al. Near-atomic resolution visualization of human transcription promoter opening. Nature 533, 359–365 (2016).
Horn, A.E., Kugel, J.F. & Goodrich, J.A. Single molecule microscopy reveals mechanistic insight into RNA polymerase II preinitiation complex assembly and transcriptional activity. Nucleic Acids Res. 44, 7132–7143 (2016).
Kostrewa, D. et al. RNA polymerase II–TFIIB structure and mechanism of transcription initiation. Nature 462, 323–330 (2009).
Rhee, H.S. & Pugh, B.F. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature 483, 295–301 (2012).
Plaschka, C. et al. Transcription initiation complex structures elucidate DNA opening. Nature 533, 353–358 (2016).
Burley, S.K. & Roeder, R.G. Biochemistry and structural biology of transcription factor IID (TFIID). Annu. Rev. Biochem. 65, 769–799 (1996).
Louder, R.K. et al. Structure of promoter-bound TFIID and model of human pre-initiation complex assembly. Nature 531, 604–609 (2016).
Verrijzer, C.P., Chen, J.L., Yokomori, K. & Tjian, R. Binding of TAFs to core elements directs promoter selectivity by RNA polymerase II. Cell 81, 1115–1125 (1995).
Jonkers, I., Kwak, H. & Lis, J.T. Genome-wide dynamics of Pol II elongation and its interplay with promoter proximal pausing, chromatin, and exons. eLife 3, e02407 (2014).
Chen, F., Gao, X. & Shilatifard, A. Stably paused genes revealed through inhibition of transcription initiation by the TFIIH inhibitor triptolide. Genes Dev. 29, 39–47 (2015).
Grünberg, S., Warfield, L. & Hahn, S. Architecture of the RNA polymerase II preinitiation complex and mechanism of ATP-dependent promoter opening. Nat. Struct. Mol. Biol. 19, 788–796 (2012).
Alekseev, S. et al. Transcription without XPB establishes a unified helicase-independent mechanism of promoter opening in eukaryotic gene expression. Mol. Cell 65, 504–514 (2017).
Zhang, Z. et al. Rapid dynamics of general transcription factor TFIIB binding during preinitiation complex assembly revealed by single-molecule analysis. Genes Dev. 30, 2106–2118 (2016).
Darzacq, X. et al. In vivo dynamics of RNA polymerase II transcription. Nat. Struct. Mol. Biol. 14, 796–806 (2007).
Henriques, T. et al. Stable pausing by RNA polymerase II provides an opportunity to target and integrate regulatory signals. Mol. Cell 52, 517–528 (2013).
Buckley, M.S., Kwak, H., Zipfel, W.R. & Lis, J.T. Kinetics of promoter Pol II on Hsp70 reveal stable pausing and key insights into its regulation. Genes Dev. 28, 14–19 (2014).
Chen, K. et al. A global change in RNA polymerase II pausing during the Drosophila midblastula transition. eLife 2, e00861 (2013).
Gaertner, B. et al. Poised RNA polymerase II changes over developmental time and prepares genes for future expression. Cell Reports 2, 1670–1683 (2012).
Zawel, L., Kumar, K.P. & Reinberg, D. Recycling of the general transcription factors during RNA polymerase II transcription. Genes Dev. 9, 1479–1490 (1995).
Yudkovsky, N., Ranish, J.A. & Hahn, S. A transcription reinitiation intermediate that is stabilized by activator. Nature 408, 225–229 (2000).
Van Dyke, M.W., Sawadogo, M. & Roeder, R.G. Stability of transcription complexes on class II genes. Mol. Cell. Biol. 9, 342–344 (1989).
Chao, S.H. & Price, D.H. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J. Biol. Chem. 276, 31793–31799 (2001).
Marshall, N.F. & Price, D.H. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J. Biol. Chem. 270, 12335–12338 (1995).
Saeki, H. & Svejstrup, J.Q. Stability, flexibility, and dynamic interactions of colliding RNA polymerase II elongation complexes. Mol. Cell 35, 191–205 (2009).
Ehrensberger, A.H., Kelly, G.P. & Svejstrup, J.Q. Mechanistic interpretation of promoter-proximal peaks and RNAPII density maps. Cell 154, 713–715 (2013).
Bothma, J.P. et al. Dynamic regulation of eve stripe 2 expression reveals transcriptional bursts in living Drosophila embryos. Proc. Natl. Acad. Sci. USA 111, 10598–10603 (2014).
Fukaya, T., Lim, B. & Levine, M. Enhancer control of transcriptional bursting. Cell 166, 358–368 (2016).
Suter, D.M. et al. Mammalian genes are transcribed with widely different bursting kinetics. Science 332, 472–474 (2011).
Zoller, B., Nicolas, D., Molina, N. & Naef, F. Structure of silent transcription intervals and noise characteristics of mammalian genes. Mol. Syst. Biol. 11, 823 (2015).
Gilchrist, D.A. et al. NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes Dev. 22, 1921–1933 (2008).
Deal, R.B., Henikoff, J.G. & Henikoff, S. Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones. Science 328, 1161–1164 (2010).
Pedraza, J.M. & Paulsson, J. Effects of molecular memory and bursting on fluctuations in gene expression. Science 319, 339–343 (2008).
Coulon, A., Chow, C.C., Singer, R.H. & Larson, D.R. Eukaryotic transcriptional dynamics: from single molecules to cell populations. Nat. Rev. Genet. 14, 572–584 (2013).
Ghavi-Helm, Y. et al. Enhancer loops appear stable during development and are associated with paused polymerase. Nature 512, 96–100 (2014).
Pettersen, E.F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Acknowledgements
We thank P. Verrijzer (Erasmus University Medical Center) for TAF2 antibodies, J. Kadonaga (University of California, San Diego) for TFIIA, TFIIB, TBP and TFIIF antibodies, J. Johnston for help with data analysis, and R. Krumlauf, R. Conaway, J. Conaway, C. Kaplan and R. Fropf for comments on the manuscript. The work was funded by the Stowers Institute for Medical Research.
Author information
Authors and Affiliations
Contributions
W.S. and J.Z. conceived the project, W.S. performed all experiments and computational analyses, and W.S. and J.Z. interpreted the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 and Supplementary Tables 2–4 (PDF 9955 kb)
Supplementary Table 1
Summary of paused Pol II half-lives across the genome. (XLS 718 kb)
Rights and permissions
About this article
Cite this article
Shao, W., Zeitlinger, J. Paused RNA polymerase II inhibits new transcriptional initiation. Nat Genet 49, 1045–1051 (2017). https://doi.org/10.1038/ng.3867
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3867
This article is cited by
-
Rapid unleashing of macrophage efferocytic capacity via transcriptional pause release
Nature (2024)
-
PRO-IP-seq tracks molecular modifications of engaged Pol II complexes at nucleotide resolution
Nature Communications (2023)
-
RNA polymerase II dynamics shape enhancer–promoter interactions
Nature Genetics (2023)
-
Lola-I is a promoter pioneer factor that establishes de novo Pol II pausing during development
Nature Communications (2023)
-
The NELF pausing checkpoint mediates the functional divergence of Cdk9
Nature Communications (2023)