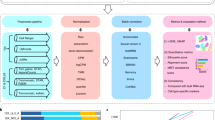
Abstract
Intratumoral heterogeneity is a major obstacle to cancer treatment and a significant confounding factor in bulk-tumor profiling. We performed an unbiased analysis of transcriptional heterogeneity in colorectal tumors and their microenvironments using single-cell RNA–seq from 11 primary colorectal tumors and matched normal mucosa. To robustly cluster single-cell transcriptomes, we developed reference component analysis (RCA), an algorithm that substantially improves clustering accuracy. Using RCA, we identified two distinct subtypes of cancer-associated fibroblasts (CAFs). Additionally, epithelial–mesenchymal transition (EMT)-related genes were found to be upregulated only in the CAF subpopulation of tumor samples. Notably, colorectal tumors previously assigned to a single subtype on the basis of bulk transcriptomics could be divided into subgroups with divergent survival probability by using single-cell signatures, thus underscoring the prognostic value of our approach. Overall, our results demonstrate that unbiased single-cell RNA–seq profiling of tumor and matched normal samples provides a unique opportunity to characterize aberrant cell states within a tumor.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Change history
12 November 2018
In the version of the article published, the author list is not accurate. Igor Cima and Min-Han Tan should have been authors, appearing after Mark Wong in the author list, while Paul Jongjoon Choi should not have been listed as an author. Igor Cima and Min-Han Tan vboth have the affiliation Institute of Bioengineering and Nanotechnology, Singapore, Singapore, and their contributions should have been noted in the Author Contributions section as "I.C. preprocessed Primary Cell Atlas data with inputs from M.-H.T." The following description of the contribution of Paul Jongjoon Choi should not have appeared: "P.J.C. supported the smFISH experiments.” In the 'RCA: global panel' section of the Online Methods, the following sentence should have appeared as the second sentence, "An expression atlas of human primary cells (the Primary Cell Atlas) was preprocessed similarly to in ref. 55," with new reference 55 (Cima, I. et al. Tumor-derived circulating endothelial cell clusters in colorectal cancer. Science Transl. Med. 8, 345ra89, 2016).
19 December 2022
A Correction to this paper has been published: https://doi.org/10.1038/s41588-022-01281-y
References
Burrell, R.A., McGranahan, N., Bartek, J. & Swanton, C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature 501, 338–345 (2013).
Meacham, C.E. & Morrison, S.J. Tumour heterogeneity and cancer cell plasticity. Nature 501, 328–337 (2013).
Hanahan, D. & Coussens, L.M. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell 21, 309–322 (2012).
Navin, N.E. The first five years of single-cell cancer genomics and beyond. Genome Res. 25, 1499–1507 (2015).
Patel, A.P. et al. Single-cell RNA–seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344, 1396–1401 (2014).
Tirosh, I. et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA–seq. Science 352, 189–196 (2016).
Treutlein, B. et al. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA–seq. Nature 509, 371–375 (2014).
Grün, D. et al. Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature 525, 251–255 (2015).
Grün, D. et al. De novo prediction of stem cell identity using single-cell transcriptome data. Cell Stem Cell 19, 266–277 (2016).
Zeisel, A. et al. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA–seq. Science 347, 1138–1142 (2015).
Satija, R., Farrell, J.A., Gennert, D., Schier, A.F. & Regev, A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502 (2015).
Hicks, S.C., Teng, M. & Irizarry, R.A. On the widespread and critical impact of systematic bias and batch effects in single-cell RNA–seq data. Preprint at bioRxiv https://doi.org/10.1101/025528 (2015).
Finak, G. et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 16, 278 (2015).
Torre, L.A. et al. Global cancer statistics, 2012. CA Cancer J. Clin. 65, 87–108 (2015).
Junttila, M.R. & de Sauvage, F.J. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature 501, 346–354 (2013).
Isella, C. et al. Stromal contribution to the colorectal cancer transcriptome. Nat. Genet. 47, 312–319 (2015).
Calon, A. et al. Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat. Genet. 47, 320–329 (2015).
Brennecke, P. et al. Single-cell transcriptome analysis reveals coordinated ectopic gene-expression patterns in medullary thymic epithelial cells. Nat. Immunol. 16, 933–941 (2015).
Chelly, J., Concordet, J.-P., Kaplan, J.-C. & Kahn, A. Illegitimate transcription: transcription of any gene in any cell type. Proc. Natl. Acad. Sci. USA 86, 2617–2621 (1989).
Eisen, M.B., Spellman, P.T., Brown, P.O. & Botstein, D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95, 14863–14868 (1998).
Hubert, L. & Arabie, P. Comparing partitions. J. Classif. 2, 193–218 (1985).
Wang, C., Zhan, X., Liang, L., Abecasis, G.R. & Lin, X. Improved ancestry estimation for both genotyping and sequencing data using projection Procrustes analysis and genotype imputation. Am. J. Hum. Genet. 96, 926–937 (2015).
Dalerba, P. et al. Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nat. Biotechnol. 29, 1120–1127 (2011).
Jung, P. et al. Isolation of human colon stem cells using surface expression of PTK7. Stem Cell Rep. 5, 979–987 (2015).
Merlos-Suárez, A. et al. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell 8, 511–524 (2011).
Cline, M.S. et al. Exploring TCGA pan-cancer data at the UCSC cancer genomics browser. Sci. Rep. 3, 2652 (2013).
Krämer, A., Green, J., Pollard, J. & Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 30, 523–530 (2014).
Randhawa, H. et al. Overexpression of peptide deformylase in breast, colon, and lung cancers. BMC Cancer 13, 321 (2013).
Peng, B. et al. Peptide deformylase inhibitor actinonin reduces celastrol's HSP70 induction while synergizing proliferation inhibition in tumor cells. BMC Cancer 14, 146 (2014).
Qian, B.-Z. & Pollard, J.W. Macrophage diversity enhances tumor progression and metastasis. Cell 141, 39–51 (2010).
Evens, A.M. Motexafin gadolinium: a redox-active tumor selective agent for the treatment of cancer. Curr. Opin. Oncol. 16, 576–580 (2004).
Hirata, A. et al. Dose-dependent roles for canonical Wnt signalling in de novo crypt formation and cell cycle properties of the colonic epithelium. Development 140, 66–75 (2013).
De Sousa, E. et al. Poor-prognosis colon cancer is defined by a molecularly distinct subtype and develops from serrated precursor lesions. Nat. Med. 19, 614–618 (2013).
Sadanandam, A. et al. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat. Med. 19, 619–625 (2013).
Marisa, L. et al. Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS Med. 10, e1001453 (2013).
Sadanandam, A. et al. Reconciliation of classification systems defining molecular subtypes of colorectal cancer: interrelationships and clinical implications. Cell Cycle 13, 353–357 (2014).
Gentles, A.J. et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat. Med. 21, 938–945 (2015).
Guinney, J. et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 21, 1350–1356 (2015).
Henry, L.R. et al. Clinical implications of fibroblast activation protein in patients with colon cancer. Clin. Cancer Res. 13, 1736–1741 (2007).
Tsujino, T. et al. Stromal myofibroblasts predict disease recurrence for colorectal cancer. Clin. Cancer Res. 13, 2082–2090 (2007).
Cobb, J.P. et al. Application of genome-wide expression analysis to human health and disease. Proc. Natl. Acad. Sci. USA 102, 4801–4806 (2005).
Calon, A. et al. Dependency of colorectal cancer on a TGF-β-driven program in stromal cells for metastasis initiation. Cancer Cell 22, 571–584 (2012).
Hawinkels, L. et al. Interaction with colon cancer cells hyperactivates TGF-β signaling in cancer-associated fibroblasts. Oncogene 33, 97–107 (2014).
Herrera, M. et al. Functional heterogeneity of cancer-associated fibroblasts from human colon tumors shows specific prognostic gene expression signature. Clin. Cancer Res. 19, 5914–5926 (2013).
Augsten, M. Cancer-associated fibroblasts as another polarized cell type of the tumor microenvironment. Front. Oncol. 4, 62 (2014).
Neri, S. et al. Cancer cell invasion driven by extracellular matrix remodeling is dependent on the properties of cancer-associated fibroblasts. J. Cancer Res. Clin. 142, 437–446 (2016).
Kakarla, S., Song, X. & Gottschalk, S. Cancer-associated fibroblasts as targets for immunotherapy. Immunother. 4, 1129–1138 (2012).
Brennen, W., Isaacs, J. & Denmeade, S. Rationale behind targeting fibroblast activation protein–expressing carcinoma-associated fibroblasts as a novel chemotherapeutic strategy. Mol. Cancer Ther. 11, 257–266 (2012).
Yang, J. et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 117, 927–939 (2004).
Fischer, K.R. et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature 527, 472–476 (2015).
Zheng, X. et al. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature 527, 525–530 (2015).
Vellinga, T. et al. Collagen-rich stroma in aggressive colon tumors induces mesenchymal gene expression and tumor cell invasion. Oncogene 35, 5263–5271 (2016).
Avery, S. et al. BCL-XL mediates the strong selective advantage of a 20q11. 21 amplification commonly found in human embryonic stem cell cultures. Stem Cell Rep. 1, 379–386 (2013).
Sengupta, D., Rayan, N.A., Lim, M., Lim, B. & Prabhakar, S. Fast, scalable and accurate differential expression analysis for single cells. Preprint at bioRxiv https://doi.org/10.1101/049734 (2016).
Acknowledgements
We would like to thank L. Suteja, C. Kang, S. Sudhagar, J. Sheik and M.N. Ramalingam for technical assistance, A. Brichkina (Institute of Molecular and Cell Biology, A*STAR) for providing the antibody to SMA, V. Sivakamasundari for guidance on single-cell protocols, M.H. Chew, R. Ten, W.J. Lim, J.H. Lai, C.Y. Ng and D. Koh for assistance with clinical sample collection, and Y. Hu, S. Ghosh, H. Kitano and D. Tan for feedback and scientific discussions. This study was supported by core funds from the Agency for Science, Technology and Research (A*STAR) and also by grant JCO1331CFG080 from A*STAR's Joint Council Office. P.R. acknowledges support from Agency of Science, Technology and Research grants IAF111091 and IAF111128 and associated in-kind contributions from industry partners Fluidigm Singapore and Becton Dickinson Holdings, respectively. S.L.K. and A.M.H. acknowledge the Strategic Positioning Fund (SPF2012/003) from the Biomedical Research Council (BMRC).
Author information
Authors and Affiliations
Contributions
H.L., E.T.C., L.J.K.W. I.B.T., P.R. and S.P. conceived the idea and designed the study. H.L. developed the computational algorithms and performed the bioinformatic analysis. E.T.C. optimized and conducted the experiments. D.S. assisted with the data analysis. Y.T. assisted with the experiments. J.J.L.G. and K.H.C. performed the smFISH experiments. S.L.K. assisted with the initial protocol validation. W.S.T. and L.K.H. extracted and preprocessed clinical samples and guided patient selection. C.C. coordinated the clinical sample registration, preprocessing and logistics. P.J.C. supported the smFISH experiments. A.H. provided guidance in experimental protocol design. H.L. and E.T.C. analyzed the data and interpreted the results. S.P. guided the development of computational algorithms. I.B.T., P.R. and S.P. provided guidance in data analysis and interpretation of the results. H.L., E.T.C., I.B.T., P.R. and S.P. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–16, Supplementary Tables 2 and 6, and Supplementary Note. (PDF 32896 kb)
Supplementary Table 1
Cell type annotation for the melanoma data set. (CSV 302 kb)
Supplementary Table 3
Differentially expressed genes between epithelial subtypes in normal mucosa. (XLSX 29 kb)
Supplementary Table 4
Cell-type-specific differentially expression analysis between normal mucosa and tumor. (XLSX 2195 kb)
Supplementary Table 5
Differentially expressed genes between bulk tumor and matched normal tissue (TCGA data). (CSV 2359 kb)
Rights and permissions
About this article
Cite this article
Li, H., Courtois, E., Sengupta, D. et al. Reference component analysis of single-cell transcriptomes elucidates cellular heterogeneity in human colorectal tumors. Nat Genet 49, 708–718 (2017). https://doi.org/10.1038/ng.3818
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3818
This article is cited by
-
Systematic immune cell dysregulation and molecular subtypes revealed by single-cell RNA-seq of subjects with type 1 diabetes
Genome Medicine (2024)
-
Mitochondrial respiratory chain component NDUFA4: a promising therapeutic target for gastrointestinal cancer
Cancer Cell International (2024)
-
An exosomal strategy for targeting cancer-associated fibroblasts mediated tumors desmoplastic microenvironments
Journal of Nanobiotechnology (2024)
-
T-cell infiltration and its regulatory mechanisms in cancers: insights at single-cell resolution
Journal of Experimental & Clinical Cancer Research (2024)
-
A single cell RNAseq benchmark experiment embedding “controlled” cancer heterogeneity
Scientific Data (2024)