Abstract
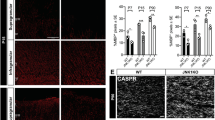
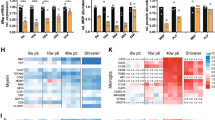
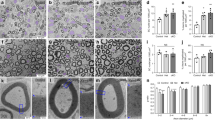
The kinesin motor protein Kif1b has previously been implicated in the axonal transport of mitochondria and synaptic vesicles1,2. More recently, KIF1B has been associated with susceptibility to multiple sclerosis (MS)3. Here we show that Kif1b is required for the localization of mbp (myelin basic protein) mRNA to processes of myelinating oligodendrocytes in zebrafish. We observe the ectopic appearance of myelin-like membrane in kif1b mutants, coincident with the ectopic localization of myelin proteins in kif1b mutant oligodendrocyte cell bodies. These observations suggest that oligodendrocytes localize certain mRNA molecules, namely those encoding small basic proteins such as MBP, to prevent aberrant effects of these proteins elsewhere in the cell. We also find that Kif1b is required for outgrowth of some of the longest axons in the peripheral and central nervous systems. Our data demonstrate previously unknown functions of kif1b in vivo and provide insights into its possible roles in MS.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Nangaku, M. et al. KIF1B, a novel microtubule plus end-directed monomeric motor protein for transport of mitochondria. Cell 79, 1209–1220 (1994).
Zhao, C. et al. Charcot-Marie-Tooth disease type 2A caused by mutation in a microtubule motor KIF1Bbeta. Cell 105, 587–597 (2001).
Aulchenko, Y.S. et al. Genetic variation in the KIF1B locus influences susceptibility to multiple sclerosis. Nat. Genet. 40, 1402–1403 (2008).
Sherman, D.L. & Brophy, P.J. Mechanisms of axon ensheathment and myelin growth. Nat. Rev. Neurosci. 6, 683–690 (2005).
Brosamle, C. & Halpern, M.E. Characterization of myelination in the developing zebrafish. Glia 39, 47–57 (2002).
Colman, D.R., Kreibich, G., Frey, A.B. & Sabatini, D.D. Synthesis and incorporation of myelin polypeptides into CNS myelin. J. Cell Biol. 95, 598–608 (1982).
Carson, J.H., Worboys, K., Ainger, K. & Barbarese, E. Translocation of myelin basic protein mRNA in oligodendrocytes requires microtubules and kinesin. Cell Motil. Cytoskeleton 38, 318–328 (1997).
Song, J., Carson, J.H., Barbarese, E., Li, F.Y. & Duncan, I.D. RNA transport in oligodendrocytes from the taiep mutant rat. Mol. Cell. Neurosci. 24, 926–938 (2003).
Ainger, K. et al. Transport and localization of exogenous myelin basic protein mRNA microinjected into oligodendrocytes. J. Cell Biol. 123, 431–441 (1993).
Pogoda, H.M. et al. A genetic screen identifies genes essential for development of myelinated axons in zebrafish. Dev. Biol. 298, 118–131 (2006).
Kikkawa, M. et al. Switch-based mechanism of kinesin motors. Nature 411, 439–445 (2001).
Marchler-Bauer, A. et al. CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res. 35, D237–D240 (2007).
Hirokawa, N. & Takemura, R. Molecular motors and mechanisms of directional transport in neurons. Nat. Rev. Neurosci. 6, 201–214 (2005).
Sato, T., Takahoko, M. & Okamoto, H. HuC:Kaede, a useful tool to label neural morphologies in networks in vivo. Genesis 44, 136–142 (2006).
Lyons, D.A., Naylor, S.G., Mercurio, S., Dominguez, C. & Talbot, W.S. KBP is essential for axonal structure, outgrowth and maintenance in zebrafish, providing insight into the cellular basis of Goldberg-Shprintzen syndrome. Development 135, 599–608 (2008).
Schlisio, S. et al. The kinesin KIF1Bbeta acts downstream from EglN3 to induce apoptosis and is a potential 1p36 tumor suppressor. Genes Dev. 22, 884–893 (2008).
Yeh, I.T. et al. A germline mutation of the KIF1B beta gene on 1p36 in a family with neural and nonneural tumors. Hum. Genet. 124, 279–285 (2008).
Munirajan, A.K. et al. KIF1Bbeta functions as a haploinsufficient tumor suppressor gene mapped to chromosome 1p36.2 by inducing apoptotic cell death. J. Biol. Chem. 283, 24426–24434 (2008).
Cahoy, J.D. et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J. Neurosci. 28, 264–278 (2008).
Shin, J., Park, H.C., Topczewska, J.M., Mawdsley, D.J. & Appel, B. Neural cell fate analysis in zebrafish using olig2 BAC transgenics. Methods Cell Sci. 25, 7–14 (2003).
Lyons, D.A. et al. erbb3 and erbb2 are essential for schwann cell migration and myelination in zebrafish. Curr. Biol. 15, 513–524 (2005).
Morris, J.K. et al. The 36K protein of zebrafish CNS myelin is a short-chain dehydrogenase. Glia 45, 378–391 (2004).
Gould, R.M., Freund, C.M. & Barbarese, E. Myelin-associated oligodendrocytic basic protein mRNAs reside at different subcellular locations. J. Neurochem. 73, 1913–1924 (1999).
Rispoli, P. et al. A thermodynamic and structural study of myelin basic protein in lipid membrane models. Biophys. J. 93, 1999–2010 (2007).
Hauser, S.L. & Oksenberg, J.R. The neurobiology of multiple sclerosis: genes, inflammation, and neurodegeneration. Neuron 52, 61–76 (2006).
Meinl, E. & Hohlfeld, R. Immunopathogenesis of multiple sclerosis: MBP and beyond. Clin. Exp. Immunol. 128, 395–397 (2002).
Hedegaard, C.J. et al. T helper cell type 1 (Th1), Th2 and Th17 responses to myelin basic protein and disease activity in multiple sclerosis. Immunology 125, 161–169 (2008).
Lambracht-Washington, D. et al. Antigen specificity of clonally expanded and receptor edited cerebrospinal fluid B cells from patients with relapsing remitting MS. J. Neuroimmunol. 186, 164–176 (2007).
Acknowledgements
We thank I. Middendorf, T. Reyes and C. Hill for technical support and fish care. We thank H. Okamoto, B. Appel, G. Jeserich and M. Meyer for sharing reagents. We thank B. Barres, P. Brophy and members of our laboratory for comments on the manuscript. This work was supported by grants from the US National Institutes of Health (NS050223, W.S.T.) and the National Multiple Sclerosis Society (RG 3943-A-2, W.S.T.), a postdoctoral fellowship from the Muscular Dystrophy Association (MDA4061, D.A.L.) and a David Phillips Fellowship from the Biotechnology and Biological Sciences Research Council, UK (D.A.L.).
Author information
Authors and Affiliations
Contributions
W.S.T. and D.A.L. designed the study. D.A.L. performed the phenotypic assessment of mutant, morphant and chimeric zebrafish, with contributions from A.S. S.G.N. mapped the st43 mutation to kif1b. D.A.L. and W.S.T. wrote this paper.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figure 1, Supplementary Methods and Supplementary Table 1 (PDF 468 kb)
Rights and permissions
About this article
Cite this article
Lyons, D., Naylor, S., Scholze, A. et al. Kif1b is essential for mRNA localization in oligodendrocytes and development of myelinated axons. Nat Genet 41, 854–858 (2009). https://doi.org/10.1038/ng.376
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.376
This article is cited by
-
Circ_0003611 regulates apoptosis and oxidative stress injury of Alzheimer’s disease via miR-383-5p/KIF1B axis
Metabolic Brain Disease (2022)
-
Cyfip1 haploinsufficient rats show white matter changes, myelin thinning, abnormal oligodendrocytes and behavioural inflexibility
Nature Communications (2019)
-
Missing in Action: Dysfunctional RNA Metabolism in Oligodendroglial Cells as a Contributor to Neurodegenerative Diseases?
Neurochemical Research (2019)
-
KIF2A characterization after spinal cord injury
Cellular and Molecular Life Sciences (2019)
-
Comprehensive analysis of coding-lncRNA gene co-expression network uncovers conserved functional lncRNAs in zebrafish
BMC Genomics (2018)