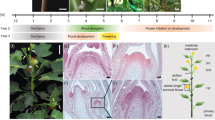
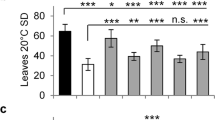
Abstract
Plants evolved so that their flowering is triggered by seasonal changes in day length1. However, day-length sensitivity in crops limits their geographical range of cultivation, and thus modification of the photoperiod response was critical for their domestication2,3,4,5,6,7,8,9,10,11. Here we show that loss of day-length-sensitive flowering in tomato was driven by the florigen paralog and flowering repressor SELF-PRUNING 5G (SP5G). SP5G expression is induced to high levels during long days in wild species, but not in cultivated tomato because of cis-regulatory variation. CRISPR/Cas9-engineered mutations in SP5G cause rapid flowering and enhance the compact determinate growth habit of field tomatoes, resulting in a quick burst of flower production that translates to an early yield. Our findings suggest that pre-existing variation in SP5G facilitated the expansion of cultivated tomato beyond its origin near the equator in South America, and they provide a compelling demonstration of the power of gene editing to rapidly improve yield traits in crop breeding.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Andrés, F. & Coupland, G. The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 13, 627–639 (2012).
Xu, M. et al. Genetic variation in four maturity genes affects photoperiod insensitivity and PHYA-regulated post-flowering responses of soybean. BMC Plant Biol. 13, 91 (2013).
Wang, Y. et al. Molecular and geographic evolutionary support for the essential role of GIGANTEAa in soybean domestication of flowering time. BMC Evol. Biol. 16, 79 (2016).
Xue, W. et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 40, 761–767 (2008).
Turner, A., Beales, J., Faure, S., Dunford, R.P. & Laurie, D.A. The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 310, 1031–1034 (2005).
Kloosterman, B. et al. Naturally occurring allele diversity allows potato cultivation in northern latitudes. Nature 495, 246–250 (2013).
Ogiso-Tanaka, E. et al. Natural variation of the RICE FLOWERING LOCUS T 1 contributes to flowering time divergence in rice. PLoS One 8, e75959 (2013).
Takahashi, Y., Teshima, K.M., Yokoi, S., Innan, H. & Shimamoto, K. Variations in Hd1 proteins, Hd3a promoters, and Ehd1 expression levels contribute to diversity of flowering time in cultivated rice. Proc. Natl. Acad. Sci. USA 106, 4555–4560 (2009).
Comadran, J. et al. Natural variation in a homolog of Antirrhinum CENTRORADIALIS contributed to spring growth habit and environmental adaptation in cultivated barley. Nat. Genet. 44, 1388–1392 (2012).
Iwata, H. et al. The TFL1 homologue KSN is a regulator of continuous flowering in rose and strawberry. Plant J. 69, 116–125 (2012).
Blackman, B.K., Strasburg, J.L., Raduski, A.R., Michaels, S.D. & Rieseberg, L.H. The role of recently derived FT paralogs in sunflower domestication. Curr. Biol. 20, 629–635 (2010).
Sawa, M., Nusinow, D.A., Kay, S.A. & Imaizumi, T. FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318, 261–265 (2007).
Suárez-López, P. et al. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410, 1116–1120 (2001).
Valverde, F. et al. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303, 1003–1006 (2004).
Tiwari, S.B. et al. The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytol. 187, 57–66 (2010).
Izawa, T. et al. Phytochrome mediates the external light signal to repress FT orthologs in photoperiodic flowering of rice. Genes Dev. 16, 2006–2020 (2002).
Ishikawa, R. et al. Phytochrome B regulates Heading date 1 (Hd1)-mediated expression of rice florigen Hd3a and critical day length in rice. Mol. Genet. Genomics 285, 461–470 (2011).
Pnueli, L. et al. The SELF-PRUNING gene of tomato regulates vegetative to reproductive switching of sympodial meristems and is the ortholog of CEN and TFL1. Development 125, 1979–1989 (1998).
Pnueli, L. et al. Tomato SP-interacting proteins define a conserved signaling system that regulates shoot architecture and flowering. Plant Cell 13, 2687–2702 (2001).
Wittwer, S.H. Photoperiod and flowering in Tomato (Lycopersicon esculentum Mill.). Proc. Am. Soc. Hortic. Sci. 83, 688–694 (1963).
Binchy, A. & Morgan, J.V. Influence of light intensity and photoperiod on inflorescence initiation in tomatoes. Isr. J. Agric. Res. 9, 261–269 (1970).
Hurd, R.G. Long-day effects on growth and flower initiation of tomato plants in low light. Ann. Appl. Biol. 73, 221–228 (1973).
Peralta, I.E. & Spooner, D.M. Morphological characterization and relationships of wild tomatoes (Solanum L. sect. Lycopersicon). Mono. Syst. Bot. 104, 227–257 (2005).
Ranc, N., Muños, S., Santoni, S. & Causse, M. A clarified position for Solanum lycopersicum var. cerasiforme in the evolutionary history of tomatoes (solanaceae). BMC Plant Biol. 8, 130 (2008).
Pease, J.B., Haak, D.C., Hahn, M.W. & Moyle, L.C. Phylogenomics reveals three sources of adaptive variation during a rapid radiation. PLoS Biol. 14, e1002379 (2016).
Takagi, H. et al. QTL-seq: rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J. 74, 174–183 (2013).
Lifschitz, E. et al. The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc. Natl. Acad. Sci. USA 103, 6398–6403 (2006).
Shalit, A. et al. The flowering hormone florigen functions as a general systemic regulator of growth and termination. Proc. Natl. Acad. Sci. USA 106, 8392–8397 (2009).
Krieger, U., Lippman, Z.B. & Zamir, D. The flowering gene SINGLE FLOWER TRUSS drives heterosis for yield in tomato. Nat. Genet. 42, 459–463 (2010).
Jiang, K., Liberatore, K.L., Park, S.J., Alvarez, J.P. & Lippman, Z.B. Tomato yield heterosis is triggered by a dosage sensitivity of the florigen pathway that fine-tunes shoot architecture. PLoS Genet. 9, e1004043 (2013).
Park, S.J. et al. Optimization of crop productivity in tomato using induced mutations in the florigen pathway. Nat. Genet. 46, 1337–1342 (2014).
Lifschitz, E., Ayre, B.G. & Eshed, Y. Florigen and anti-florigen—a systemic mechanism for coordinating growth and termination in flowering plants. Front. Plant Sci. 5, 465 (2014).
Cao, K. et al. Four tomato FLOWERING LOCUS T-like proteins act antagonistically to regulate floral initiation. Front. Plant Sci. 6, 1213 (2016).
Hanzawa, Y., Money, T. & Bradley, D. A single amino acid converts a repressor to an activator of flowering. Proc. Natl. Acad. Sci. USA 102, 7748–7753 (2005).
Ahn, J.H. et al. A divergent external loop confers antagonistic activity on floral regulators FT and TFL1. EMBO J. 25, 605–614 (2006).
Pin, P.A. et al. An antagonistic pair of FT homologs mediates the control of flowering time in sugar beet. Science 330, 1397–1400 (2010).
Ho, W.W.H. & Weigel, D. Structural features determining flower-promoting activity of Arabidopsis FLOWERING LOCUS T. Plant Cell 26, 552–564 (2014).
Eshed, Y. & Zamir, D. An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield-associated QTL. Genetics 141, 1147–1162 (1995).
Bolger, A. et al. The genome of the stress-tolerant wild tomato species Solanum pennellii. Nat. Genet. 46, 1034–1038 (2014).
deVicente, M.C. & Tanksley, S.D. QTL analysis of transgressive segregation in an interspecific tomato cross. Genetics 134, 585–596 (1993).
Jones, C.M., Rick, C.M., Adams, D., Jernstedt, J. & Chetelat, R.T. Genealogy and fine mapping of obscuravenosa, a gene affecting the distribution of chloroplasts in leaf veins, and evidence of selection during breeding of tomatoes (Lycopersicon esculentum; Solanaceae). Am. J. Bot. 94, 935–947 (2007).
Chitwood, D.H. et al. A quantitative genetic basis for leaf morphology in a set of precisely defined tomato introgression lines. Plant Cell 25, 2465–2481 (2013).
Birchler, J.A. & Veitia, R.A. The gene balance hypothesis: implications for gene regulation, quantitative traits and evolution. New Phytol. 186, 54–62 (2010).
Park, S.J., Jiang, K., Schatz, M.C. & Lippman, Z.B. Rate of meristem maturation determines inflorescence architecture in tomato. Proc. Natl. Acad. Sci. USA 109, 639–644 (2012).
Tomato Genome Consortium. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641 (2012).
Strickler, S.R. et al. Comparative genomics and phylogenetic discordance of cultivated tomato and close wild relatives. PeerJ 3, e793 (2015).
Liu, L. et al. Induced and natural variation of promoter length modulates the photoperiodic response of FLOWERING LOCUS T. Nat. Commun. 5, 4558 (2014).
Doudna, J.A. & Charpentier, E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346, 1258096 (2014).
Belhaj, K., Chaparro-Garcia, A., Kamoun, S., Patron, N.J. & Nekrasov, V. Editing plant genomes with CRISPR/Cas9. Curr. Opin. Biotechnol. 32, 76–84 (2015).
Brooks, C., Nekrasov, V., Lippman, Z.B. & Van Eck, J. Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated9 system. Plant Physiol. 166, 1292–1297 (2014).
Thouet, J., Quinet, M., Ormenese, S., Kinet, J.-M. & Périlleux, C. Revisiting the involvement of SELF-PRUNING in the sympodial growth of tomato. Plant Physiol. 148, 61–64 (2008).
Yeager, A.F. Determinate growth in the tomato. J. Hered. 18, 263–266 (1927).
MacArthur, J.W. Inherited characters in tomato. I—The self pruning habit. J. Hered. 23, 394–395 (1932).
Abelenda, J.A., Cruz-Oró, E., Franco-Zorrilla, J.M. & Prat, S. Potato StCONSTANS-like1 suppresses storage organ formation by directly activating the FT-like StSP5G repressor. Curr. Biol. 26, 872–881 (2016).
Taoka, K. et al. 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 476, 332–335 (2011).
Jaeger, K.E., Pullen, N., Lamzin, S., Morris, R.J. & Wigge, P.A. Interlocking feedback loops govern the dynamic behavior of the floral transition in Arabidopsis. Plant Cell 25, 820–833 (2013).
Rick, C.M. The Tomato. Sci. Am. 239, 76–87 (1978).
Müller, N.A. et al. Domestication selected for deceleration of the circadian clock in cultivated tomato. Nat. Genet. 48, 89–93 (2016).
Meng, X., Muszynski, M.G. & Danilevskaya, O.N. The FT-like ZCN8 gene functions as a floral activator and is involved in photoperiod sensitivity in maize. Plant Cell 23, 942–960 (2011).
Hecht, V. et al. The pea GIGAS gene is a FLOWERING LOCUS T homolog necessary for graft-transmissible specification of flowering but not for responsiveness to photoperiod. Plant Cell 23, 147–161 (2011).
Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013).
R Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2014).
Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013).
Werner, S., Engler, C., Weber, E., Gruetzner, R. & Marillonnet, S. Fast track assembly of multigene constructs using Golden Gate cloning and the MoClo system. Bioeng. Bugs 3, 38–43 (2012).
Van Eck, J., Kirk, D.D. & Walmsley, A.M. in Agrobacterium Protocols (ed. Wang, K.) 459–473 (Humana Press Inc., 2006).
Acknowledgements
We thank all members of the Lippman lab and Y. Eshed for valuable discussions. We also thank M. Koornneef for his constant support and useful discussions. We thank C. Brooks, A. Krainer and J. Dalrymple for technical support; T. Mulligan; S. Vermylen; A. Krainer from CSHL; K. Dunn and M. Treat from Robert Treat Farm in Milford, Connecticut; and staff from Cornell University's Long Island Horticultural Research and Extension Center in Riverhead, New York, for assistance with plant care. We thank U. Tartler and A. Lautscham for their help at the Max Planck Institute for Plant Breeding Research. This research was supported by an EMBO Long-Term Fellowship (ALTF 1589-2014 to S.S.), the Next-Generation BioGreen 21 Program (PMBC, PJ011912012016 to S.J.P.), the German Research Foundation (DFG project number SCHM2793/1-1 to I.S.), the Max Planck Society (I.S.), the German Research Foundation under the German-Israeli Project Cooperation program (DFG DIP project number FE552/12-1 to J.M.J.-G.), the National Science Foundation Plant Genome Research Program (IOS-1237880 to J.V.E. and Z.B.L.), BARD (IS-4818-15 to Z.B.L.), the US-Israel Binational Agricultural Research & Development fund (Z.B.L.), and an Agriculture and Food Research Initiative competitive grant from the USDA National Institute of Food and Agriculture (2016-67013-24452 to Z.B.L.).
Author information
Authors and Affiliations
Contributions
S.S., N.A.M., S.J.P., I.S., K.J., R.H., L.Z., J.V.E., J.M.J.-G. and Z.B.L. designed and planned experiments. S.S., N.A.M., S.J.P., I.S., K.J., R.H., L.Z., J.M.J.-G. and Z.B.L. performed experiments and collected the data. S.S., N.A.M., S.J.P., I.S., K.J., R.H., J.M.J.-G. and Z.B.L. analyzed the data. S.S., N.A.M., J.M.J.-G. and Z.B.L designed the research. S.S. and Z.B.L. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
S.S., S.J.P. and Z.B.L. have filed a PCT patent application based in part on this work (US Provisional Application No. 62/320,439) with the US Patent and Trademark Office.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–15 and Supplementary Tables 1–5 (PDF 5633 kb)
Supplementary Table
Source data for Supplementary Figure 14 (XLSX 25 kb)
Rights and permissions
About this article
Cite this article
Soyk, S., Müller, N., Park, S. et al. Variation in the flowering gene SELF PRUNING 5G promotes day-neutrality and early yield in tomato. Nat Genet 49, 162–168 (2017). https://doi.org/10.1038/ng.3733
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3733
This article is cited by
-
Precise fine-turning of GhTFL1 by base editing tools defines ideal cotton plant architecture
Genome Biology (2024)
-
Comparative yield evaluation of mini-tomato cultivar in two hydroponic systems
Horticulture, Environment, and Biotechnology (2024)
-
Genome editing in cotton: challenges and opportunities
Journal of Cotton Research (2023)
-
Genetic architecture of fresh-market tomato yield
BMC Plant Biology (2023)
-
Tight genetic linkage of genes causing hybrid necrosis and pollinator isolation between young species
Nature Plants (2023)