Abstract
There is a striking and unexplained male predominance across many cancer types. A subset of X-chromosome genes can escape X-inactivation, which would protect females from complete functional loss by a single mutation. To identify putative 'escape from X-inactivation tumor-suppressor' (EXITS) genes, we examined somatic alterations from >4,100 cancers across 21 tumor types for sex bias. Six of 783 non-pseudoautosomal region (PAR) X-chromosome genes (ATRX, CNKSR2, DDX3X, KDM5C, KDM6A, and MAGEC3) harbored loss-of-function mutations more frequently in males (based on a false discovery rate < 0.1), in comparison to zero of 18,055 autosomal and PAR genes (Fisher's exact P < 0.0001). Male-biased mutations in genes that escape X-inactivation were observed in combined analysis across many cancers and in several individual tumor types, suggesting a generalized phenomenon. We conclude that biallelic expression of EXITS genes in females explains a portion of the reduced cancer incidence in females as compared to males across a variety of tumor types.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
References
Edgren, G., Liang, L., Adami, H.O. & Chang, E.T. Enigmatic sex disparities in cancer incidence. Eur. J. Epidemiol. 27, 187–196 (2012).
Cook, M.B. et al. Sex disparities in cancer incidence by period and age. Cancer Epidemiol. Biomarkers Prev. 18, 1174–1182 (2009).
Siegel, R., Naishadham, D. & Jemal, A. Cancer statistics, 2013. CA Cancer J. Clin. 63, 11–30 (2013).
Van der Meulen, J. et al. The H3K27me3 demethylase UTX is a gender-specific tumor suppressor in T-cell acute lymphoblastic leukemia. Blood 125, 13–21 (2015).
Yoshida, K. et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 478, 64–69 (2011).
Van Vlierberghe, P. et al. PHF6 mutations in T-cell acute lymphoblastic leukemia. Nat. Genet. 42, 338–342 (2010).
van Haaften, G. et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat. Genet. 41, 521–523 (2009).
Knudson, A.G. Antioncogenes and human cancer. Proc. Natl. Acad. Sci. USA 90, 10914–10921 (1993).
Yang, C. et al. X-chromosome inactivation: molecular mechanisms from the human perspective. Hum. Genet. 130, 175–185 (2011).
Berletch, J.B. et al. Escape from X inactivation varies in mouse tissues. PLoS Genet. 11, e1005079 (2015).
Berletch, J.B., Yang, F., Xu, J., Carrel, L. & Disteche, C.M. Genes that escape from X inactivation. Hum. Genet. 130, 237–245 (2011).
Carrel, L. & Willard, H.F. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature 434, 400–404 (2005).
Johnston, C.M. et al. Large-scale population study of human cell lines indicates that dosage compensation is virtually complete. PLoS Genet. 4, e9 (2008).
Talebizadeh, Z., Simon, S.D. & Butler, M.G. X chromosome gene expression in human tissues: male and female comparisons. Genomics 88, 675–681 (2006).
Duijf, P.H., Schultz, N. & Benezra, R. Cancer cells preferentially lose small chromosomes. Int. J. Cancer 132, 2316–2326 (2013).
Yildirim, E. et al. Xist RNA is a potent suppressor of hematologic cancer in mice. Cell 152, 727–742 (2013).
Veiga, L.C., Bérgamo, N.A., Reis, P.P., Kowalski, L.P. & Rogatto, S.R. Loss of Y-chromosome does not correlate with age at onset of head and neck carcinoma: a case–control study. Braz. J. Med. Biol. Res. 45, 172–178 (2012).
Zhang, L.J., Shin, E.S., Yu, Z.X. & Li, S.B. Molecular genetic evidence of Y chromosome loss in male patients with hematological disorders. Chin. Med. J. (Engl.) 120, 2002–2005 (2007).
Dumanski, J.P. et al. Smoking is associated with mosaic loss of chromosome Y. Science 347, 81–83 (2015).
Forsberg, L.A. et al. Mosaic loss of chromosome Y in peripheral blood is associated with shorter survival and higher risk of cancer. Nat. Genet. 46, 624–628 (2014).
Bellott, D.W. et al. Mammalian Y chromosomes retain widely expressed dosage-sensitive regulators. Nature 508, 494–499 (2014).
Cortez, D. et al. Origins and functional evolution of Y chromosomes across mammals. Nature 508, 488–493 (2014).
Shpargel, K.B., Sengoku, T., Yokoyama, S. & Magnuson, T. UTX and UTY demonstrate histone demethylase–independent function in mouse embryonic development. PLoS Genet. 8, e1002964 (2012).
Welstead, G.G. et al. X-linked H3K27me3 demethylase Utx is required for embryonic development in a sex-specific manner. Proc. Natl. Acad. Sci. USA 109, 13004–13009 (2012).
Brat, D.J. et al. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N. Engl. J. Med. 372, 2481–2498 (2015).
Pinto, E.M. et al. Genomic landscape of paediatric adrenocortical tumours. Nat. Commun. 6, 6302 (2015).
Jiang, L. et al. Exome sequencing identifies somatic mutations of DDX3X in natural killer/T-cell lymphoma. Nat. Genet. 47, 1061–1066 (2015).
Dalgliesh, G.L. et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature 463, 360–363 (2010).
Ntziachristos, P. et al. Contrasting roles of histone 3 lysine 27 demethylases in acute lymphoblastic leukaemia. Nature 514, 513–517 (2014).
Dubrow, R. & Darefsky, A.S. Demographic variation in incidence of adult glioma by subtype, United States, 1992–2007. BMC Cancer 11, 325 (2011).
Wilson Sayres, M.A. & Makova, K.D. Gene survival and death on the human Y chromosome. Mol. Biol. Evol. 30, 781–787 (2013).
Bechara, E.G., Sebestyén, E., Bernardis, I., Eyras, E. & Valcárcel, J. RBM5, 6, and 10 differentially regulate NUMB alternative splicing to control cancer cell proliferation. Mol. Cell 52, 720–733 (2013).
Cotton, A.M. et al. Analysis of expressed SNPs identifies variable extents of expression from the human inactive X chromosome. Genome Biol. 14, R122 (2013).
Al Nadaf, S. et al. A cross-species comparison of escape from X inactivation in Eutheria: implications for evolution of X chromosome inactivation. Chromosoma 121, 71–78 (2012).
Chaligné, R. et al. The inactive X chromosome is epigenetically unstable and transcriptionally labile in breast cancer. Genome Res. 25, 488–503 (2015).
Patrat, C. et al. Dynamic changes in paternal X-chromosome activity during imprinted X-chromosome inactivation in mice. Proc. Natl. Acad. Sci. USA 106, 5198–5203 (2009).
GTEx Consortium. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–660 (2015).
Wu, H. et al. Cellular resolution maps of X chromosome inactivation: implications for neural development, function, and disease. Neuron 81, 103–119 (2014).
Lawrence, M.S. et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505, 495–501 (2014).
Vaags, A.K. et al. Absent CNKSR2 causes seizures and intellectual, attention, and language deficits. Ann. Neurol. 76, 758–764 (2014).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550 (2005).
Liberzon, A. et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015).
Chaligné, R. & Heard, E. X-chromosome inactivation in development and cancer. FEBS Lett. 588, 2514–2522 (2014).
Gibbons, R.J. & Higgs, D.R. Molecular-clinical spectrum of the ATR-X syndrome. Am. J. Med. Genet. 97, 204–212 (2000).
Jensen, L.R. et al. Mutations in the JARID1C gene, which is involved in transcriptional regulation and chromatin remodeling, cause X-linked mental retardation. Am. J. Hum. Genet. 76, 227–236 (2005).
Miyake, N. et al. MLL2 and KDM6A mutations in patients with Kabuki syndrome. Am. J. Med. Genet. A. 161A, 2234–2243 (2013).
Snijders Blok, L. et al. Mutations in DDX3X are a common cause of unexplained intellectual disability with gender-specific effects on Wnt signaling. Am. J. Hum. Genet. 97, 343–352 (2015).
Pinto, D. et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature 466, 368–372 (2010).
Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427 (2014).
Zack, T.I. et al. Pan-cancer patterns of somatic copy number alteration. Nat. Genet. 45, 1134–1140 (2013).
Gao, J. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 (2013).
Cerami, E. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012).
Siepel, A. et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15, 1034–1050 (2005).
Carter, S.L. et al. Absolute quantification of somatic DNA alterations in human cancer. Nat. Biotechnol. 30, 413–421 (2012).
Beroukhim, R. et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc. Natl. Acad. Sci. USA 104, 20007–20012 (2007).
Yoda, A. et al. Mutations in G protein β subunits promote transformation and kinase inhibitor resistance. Nat. Med. 21, 71–75 (2015).
Hammerman, P.S. et al. Mutations in the DDR2 kinase gene identify a novel therapeutic target in squamous cell lung cancer. Cancer Discov. 1, 78–89 (2011).
Lane, A.A. et al. Triplication of a 21q22 region contributes to B cell transformation through HMGN1 overexpression and loss of histone H3 Lys27 trimethylation. Nat. Genet. 46, 618–623 (2014).
Acknowledgements
The authors thank C. Sievers and B. Bernstein for helpful discussions, Z. Herbert and the Molecular Biology Core Facilities at the Dana-Farber Cancer Institute for assistance with RNA–seq, and T. Golub for facilitating the project. This work was supported by National Cancer Institute grant K08CA181340 (A.A.L.), an American Society of Hematology Scholar Award (A.A.L.), and a V Foundation Scholar Award (A.A.L.). G.G. is the Paul C. Zamecnik, MD, Chair in Oncology at Massachusetts General Hospital. D.M.W. is a Leukemia and Lymphoma Society Scholar and is supported by a Stand Up to Cancer Innovative Research Grant.
Author information
Authors and Affiliations
Contributions
A.D., V.S., S.E.S., J.P.C., A.Y., T.J.S., and J.M.H. designed, performed, and analyzed the data from computational and laboratory analyses. D.M.W., A.A.G., R.B., M.S.L., G.G., and A.A.L. conceived the study, designed experiments, and interpreted the data. A.D., D.M.W., M.S.L., G.G., and A.A.L. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
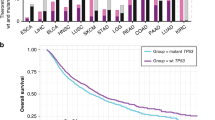
Supplementary Figure 1 SEER data for male and female cancer incidence from 1975–2013.
(a,b) Incidence rates of oral cavity and pharynx cancer (a) and leukemia (b) are plotted, grouped by males (blue), females (green), or all patients (black).
Supplementary Figure 2 Male and female mutation data across cancer types.
(a) Ratio of female:male (F:M) coding mutations across all 4,126 cancers with exome sequencing by chromosome. The F:M ratio for the X chromosome is approximately double that for autosomes, as expected because females have two X chromosomes. On average, there are more mutations in male cancers than in female cancers across all chromosomes. This finding makes our approach to discover male-cancer-biased mutations on the X chromosome even more stringent because, to be enriched, they must be present at an M:F ratio that statistically exceeds the already male-biased background rate of mutation. (b) Female (red) and male (blue) somatic mutation rates (per Mb of DNA) by cancer type. Each dot represents one cancer sample. The horizontal bar represents the median number of mutations in female or male cancers. Numbers across the top of the graph are the number of tumor–normal pairs in each disease. Disease acronym definitions are provided in Supplementary Table 1.
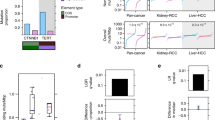
Supplementary Figure 3 Defining copy number loss events on the X chromosome.
(a) The M:F ratio of copy number loss events (y axis) in a sliding window of different lengths over the genome (x axis). The blue line represents the cutoff chosen to define focal events (Online Methods). (b) Distribution of the log2 (copy number) in tumor as compared to paired normal samples by genomic site on the X chromosome in males (top) and females (bottom). The red line represents the cutoff below which focal copy number loss was called. (c) Distribution of estimated X-chromosome ploidy across 10,844 tumors in the Broad Institute TCGA/GDAC data set (http://gdac.broadinstitute.org/) as determined by SNP array (left). The red line represents the cutoff used to infer loss of the whole X chromosome in female tumors in our data set. Estimated X-chromosome copy numbers for female and male tumors in our data set with concurrent LOF or LOF/CN loss mutations are shown on the right.
Supplementary Figure 4 Genes with higher frequencies of loss-of-function mutations in male cancers identified by a log-likelihood ratio test.
(a–c) Log-likelihood ratios for genes on the X chromosome across all 4,126 cancers (a), in clear cell kidney cancer (KIRC) (b), and in lower-grade glioma (LGG) (c) are shown. The log2 (M:F ratio) of patients with mutations is plotted against the significance value. The size and color of each circle represent the number of patients with loss-of-function mutations for each gene. Genes with FDR <0.1 for male-cancer-biased mutation are identified. ATRX is also shown, as it was identified as more frequent in male cancers by the permutation analysis (Table 1 and Fig. 2a).
Supplementary Figure 5 Male-biased mutations are more frequent among X-chromosome genes that escape X-inactivation or have functional Y-chromosome homologs.
(a) M:F ratio (left; P = 0.022 by t test; each bar represents the median; ‘+’, mean; box, interquartile range; whiskers, 10th–90th percentile) and scatterplot (right; P = 0.011 by Komolgorov–Smirnov (K–S) test; each point represents the number of males versus the number of females with a mutation in that gene; the line represents linear regression fit) of putative loss-of-function SNV, indel, or copy number loss for 46 mutated X-chromosome genes that escape X-inactivation (Escape) and the remaining 595 mutated X-chromosome genes (Non-Escape). (b) M:F ratio (left; P = 0.058 by t test; plot as in a) and scatterplot (right; P = 0.068 by K–S test) of putative LOF mutations as above for X-chromosome genes that do (Y homolog; n = 17) or do not (Non-Y homolog; n = 624) have predicted functional Y-chromosome homologs.
Supplementary Figure 6 Determination of Y-chromosome loss from exome sequencing data.
The relative number of reads between tumor and normal samples for females (red) and males (blue) on the Y chromosome is shown. The large blue dots represent samples called as having loss of Y chromosome, on the basis of having <25% of the mean number of reads among males.
Supplementary Figure 7 Allele-specific expression of EXITS genes in TCGA tumors.
Each dot represents the average minor allele expression fraction from RNA–seq at heterozygous SNP sites detected in exome sequencing in known non-escape genes on the X chromosome or EXITS genes, in tumors without mutations in the indicated gene (red cross, mean; green square, median). ‘Non-escape’ genes are the non-PAR X-chromosome genes not defined as ‘stringent’ escape genes in Supplementary Table 8. Tumor types are as follows: GBM, glioblastoma; LGG, lower-grade glioma; HNSC, head and neck squamous cell carcinoma; KIRC, clear cell kidney cancer; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma. Violin plots were generated using the distributionPlot.m function in MATLAB with default parameters.
Supplementary Figure 8 Comparison of female and male expression levels of EXITS genes in non-mutated tumors.
Female and male expression of ATRX, CNKSR2, DDX3X, KDM5C, and KDM6A in head and neck squamous cell carcinoma (HNSC), clear cell kidney cancer (KIRC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), glioblastoma (GBM), and lower-grade glioma (LGG) are shown. Analysis is limited to tumors without mutations in the indicated genes. Each bar represents the median; box, interquartile range; whiskers, 10th–90th percentile. P values were calculated by K–S test; comparisons with P < 0.05 are shaded in blue.
Supplementary Figure 9 Allele-specific expression of EXITS genes in female GTEx normal tissues.
Each dot represents the average minor allele expression fraction from RNA–seq at heterozygous SNP sites detected in exome sequencing in known non-escape genes on the X chromosome or EXITS genes, in a GTEx biopsy of the indicated normal tissue (red cross, mean; green square, median). Violin plots were generated using the distributionPlot.m function in MATLAB with default parameters.
Supplementary Figure 10 ATRX expression in male and female tissues measured by RNA sequencing.
(a) Expression of ATRX in normal male and female peripheral blood and brain in the GTEx data set is shown (distributions compared by Komolgorov–Smirnov test). (b) Holzmann–Volmer likelihood test of restricted (yellow) and unrestricted (transparent gray) normal mixture fits for males (left) and females (right). Top, maximum-likelihood fits with equal variance of the component mixtures; bottom, maximum-likelihood fits with unequal variance of the component mixtures. The P value of the Holzmann–Vollmer likelihood-ratio test for bimodality is indicated. (c) Wilcoxon rank-sum test of significance P values for difference in EXITS gene expression between males and females in the indicated tissues in the GTEx data set.
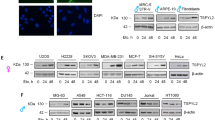
Supplementary Figure 11 Loss of Cnksr2 is associated with oncogenic phenotypes.
(a) Unsupervised hierarchical clustering of Spearman rank correlations across nine biologically independent cultures of NIH 3T3 cells stably expressing lentiviruses with the indicated shRNAs. Red–blue color represents high–low correlation, as shown in the legend. shCnksr2 1 and shCnksr2 2 are independent shRNAs, each represented in three clones. shControl is a non-targeting shRNA (targeting RFP, a control gene not present in these cells). (b) Principal-component analysis of expression profiles in the same cells as in a. (c) Network enrichment map of GSEA results querying the C2:CGP database of MSigDB, showing the interconnected cluster with the largest number of gene set members. Red circles are gene sets enriched in shCnksr2 cells and are labeled; blue circles are gene sets enriched in shControl cells. The size of each circle represents the number of genes in the gene set, and lines connect gene sets with overlapping members. (d) GSEA results querying the H1:Hallmarks database of MSigDB showing enrichment of MTORC1 and KRAS signaling pathways in shCnksr2 cells. ES, enrichment score. (e) Immunoblotting for phospho-ERK, total ERK, or tubulin in NIH 3T3 cells stably expressing the indicated shRNAs. (f) Automated counts of colonies per field of NIH 3T3 cells stably expressing the indicated shRNAs grown in soft agar (each graph represents four fields counted in two independently transfected wells; *P < 0.05 by t test as compared to shRFP; error bars, s.e.m.). (g) Representative bright-field photomicrographs of the soft agar colony assays quantified in f.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–11. (PDF 2177 kb)
Supplementary Tables 1–12
Supplementary Tables 1–12. (XLSX 1027 kb)
Rights and permissions
About this article
Cite this article
Dunford, A., Weinstock, D., Savova, V. et al. Tumor-suppressor genes that escape from X-inactivation contribute to cancer sex bias. Nat Genet 49, 10–16 (2017). https://doi.org/10.1038/ng.3726
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3726
This article is cited by
-
XX sex chromosome complement modulates immune responses to heat-killed Streptococcus pneumoniae immunization in a microbiome-dependent manner
Biology of Sex Differences (2024)
-
Sex and statin-related genetic associations at the PCSK9 gene locus: results of genome-wide association meta-analysis
Biology of Sex Differences (2024)
-
Molecular and clinical analyses of PHF6 mutant myeloid neoplasia provide their pathogenesis and therapeutic targeting
Nature Communications (2024)
-
Compartment-specific multiomic profiling identifies SRC and GNAS as candidate drivers of epithelial-to-mesenchymal transition in ovarian carcinosarcoma
British Journal of Cancer (2024)
-
Hallmarks of sex bias in immuno-oncology: mechanisms and therapeutic implications
Nature Reviews Cancer (2024)