Abstract
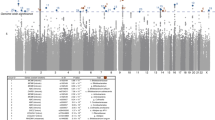
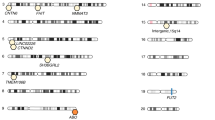
The gut microbiome is affected by multiple factors, including genetics. In this study, we assessed the influence of host genetics on microbial species, pathways and gene ontology categories, on the basis of metagenomic sequencing in 1,514 subjects. In a genome-wide analysis, we identified associations of 9 loci with microbial taxonomies and 33 loci with microbial pathways and gene ontology terms at P < 5 × 10−8. Additionally, in a targeted analysis of regions involved in complex diseases, innate and adaptive immunity, or food preferences, 32 loci were identified at the suggestive level of P < 5 × 10−6. Most of our reported associations are new, including genome-wide significance for the C-type lectin molecules CLEC4F–CD207 at 2p13.3 and CLEC4A–FAM90A1 at 12p13. We also identified association of a functional LCT SNP with the Bifidobacterium genus (P = 3.45 × 10−8) and provide evidence of a gene–diet interaction in the regulation of Bifidobacterium abundance. Our results demonstrate the importance of understanding host–microbe interactions to gain better insight into human health.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
References
Tigchelaar, E.F. et al. Gut microbiota composition associated with stool consistency. Gut 65, 540–542 (2015).
Imhann, F. et al. Proton pump inhibitors affect the gut microbiome. Gut 65, 740–748 (2015).
Zhernakova, A. et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science 352, 565–569 (2016).
David, L.A. et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563 (2014).
Scott, K.P., Gratz, S.W., Sheridan, P.O., Flint, H.J. & Duncan, S.H. The influence of diet on the gut microbiota. Pharmacol. Res. 69, 52–60 (2013).
Goodrich, J.K. et al. Human genetics shape the gut microbiome. Cell 159, 789–799 (2014).
Org, E. et al. Genetic and environmental control of host–gut microbiota interactions. Genome Res. 25, 1558–1569 (2015).
Leamy, L.J. et al. Host genetics and diet, but not immunoglobulin A expression, converge to shape compositional features of the gut microbiome in an advanced intercross population of mice. Genome Biol. 15, 552 (2014).
Goodrich, J.K. et al. Genetic determinants of the gut microbiome in UK Twins. Cell Host Microbe 19, 731–743 (2016).
Blekhman, R. et al. Host genetic variation impacts microbiome composition across human body sites. Genome Biol. 16, 191 (2015).
Davenport, E.R. et al. Genome-wide association studies of the human gut microbiota. PLoS One 10, e0140301 (2015).
Srinivas, G. et al. Genome-wide mapping of gene–microbiota interactions in susceptibility to autoimmune skin blistering. Nat. Commun. 4, 2462 (2013).
Parks, B.W. et al. Genetic control of obesity and gut microbiota composition in response to high-fat, high-sucrose diet in mice. Cell Metab. 17, 141–152 (2013).
McKnite, A.M. et al. Murine gut microbiota is defined by host genetics and modulates variation of metabolic traits. PLoS One 7, e39191 (2012).
Jostins, L. et al. Host–microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491, 119–124 (2012).
Knights, D. et al. Complex host genetics influence the microbiome in inflammatory bowel disease. Genome Med. 6, 107 (2014).
Lamas, B. et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat. Med. 22, 598–605 (2016).
Tigchelaar, E.F. et al. Cohort profile: LifeLines DEEP, a prospective, general population cohort study in the northern Netherlands: study design and baseline characteristics. BMJ Open 5, e006772 (2015).
Netea, M.G. et al. Understanding human immune function using the resources from the Human Functional Genomics Project. Nat. Med. 22, 831–833 (2016).
Mujagic, Z. et al. Small intestinal permeability is increased in diarrhoea predominant IBS, while alterations in gastroduodenal permeability in all IBS subtypes are largely attributable to confounders. Aliment. Pharmacol. Ther. 40, 288–297 (2014).
Juste, C. et al. Bacterial protein signals are associated with Crohn's disease. Gut 63, 1566–1577 (2014).
Liu, T.-C. et al. O-011 Paneth cell phenotypes define a subtype of pediatric Crohn's disease through alterations in host–microbial interactions. Inflamm. Bowel Dis. 22 (Suppl. 1), S4 (2016).
Torres, J. et al. The features of mucosa-associated microbiota in primary sclerosing cholangitis. Aliment. Pharmacol. Ther. 43, 790–801 (2016).
Hromatka, B.S. et al. Genetic variants associated with motion sickness point to roles for inner ear development, neurological processes and glucose homeostasis. Hum. Mol. Genet. 24, 2700–2708 (2015).
Locke, A.E. et al. Genetic studies of body mass index yield new insights for obesity biology. Nature 518, 197–206 (2015).
Williams, A.L. et al. Sequence variants in SLC16A11 are a common risk factor for type 2 diabetes in Mexico. Nature 506, 97–101 (2014).
Fu, J. et al. The gut microbiome contributes to a substantial proportion of the variation in blood lipids. Circ. Res. 117, 817–824 (2015).
Martínez, I. et al. Gut microbiome composition is linked to whole grain–induced immunological improvements. ISME J. 7, 269–280 (2013).
Tyler, A.D. et al. Characterization of the gut-associated microbiome in inflammatory pouch complications following ileal pouch-anal anastomosis. PLoS One 8, e66934 (2013).
Landy, J. et al. Variable alterations of the microbiota, without metabolic or immunological change, following faecal microbiota transplantation in patients with chronic pouchitis. Sci. Rep. 5, 12955 (2015).
Kalabis, J., Rosenberg, I. & Podolsky, D.K. Vangl1 protein acts as a downstream effector of intestinal trefoil factor (ITF)/TFF3 signaling and regulates wound healing of intestinal epithelium. J. Biol. Chem. 281, 6434–6441 (2006).
Bae, J.A. et al. An unconventional KITENIN/ErbB4-mediated downstream signal of EGF upregulates c-Jun and the invasiveness of colorectal cancer cells. Clin. Cancer Res. 20, 4115–4128 (2014).
Lee, S. et al. Expression of KITENIN in human colorectal cancer and its relation to tumor behavior and progression. Pathol. Int. 61, 210–220 (2011).
Klingberg, S. et al. Inverse relation between dietary intake of naturally occurring plant sterols and serum cholesterol in northern Sweden. Am. J. Clin. Nutr. 87, 993–1001 (2008).
Kaplan, R.C. et al. A genome-wide association study identifies novel loci associated with circulating IGF-I and IGFBP-3. Hum. Mol. Genet. 20, 1241–1251 (2011).
Liu, Y.J. et al. Genome-wide association scans identified CTNNBL1 as a novel gene for obesity. Hum. Mol. Genet. 17, 1803–1813 (2008).
Strawbridge, R.J. et al. Genome-wide association identifies nine common variants associated with fasting proinsulin levels and provides new insights into the pathophysiology of type 2 diabetes. Diabetes 60, 2624–2634 (2011).
Khan, M.T., Browne, W.R., van Dijl, J.M. & Harmsen, H.J.M. How can Faecalibacterium prausnitzii employ riboflavin for extracellular electron transfer? Antioxid. Redox Signal. 17, 1433–1440 (2012).
Morgan, X.C. et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 13, R79 (2012).
Lozupone, C.A., Stombaugh, J.I., Gordon, J.I., Jansson, J.K. & Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 489, 220–230 (2012).
Bønnelykke, K. et al. Meta-analysis of genome-wide association studies identifies ten loci influencing allergic sensitization. Nat. Genet. 45, 902–906 (2013).
Gevers, D. et al. The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe 15, 382–392 (2014).
Caesar, R., Tremaroli, V., Kovatcheva-Datchary, P., Cani, P.D. & Bäckhed, F. Crosstalk between gut microbiota and dietary lipids aggravates WAT inflammation through TLR signaling. Cell Metab. 22, 658–668 (2015).
Yahiro, K. et al. DAP1, a negative regulator of autophagy, controls SubAB-mediated apoptosis and autophagy. Infect. Immun. 82, 4899–4908 (2014).
Koren, I., Reem, E. & Kimchi, A. DAP1, a novel substrate of mTOR, negatively regulates autophagy. Curr. Biol. 20, 1093–1098 (2010).
Glocker, E.O. et al. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N. Engl. J. Med. 361, 2033–2045 (2009).
Galeone, M., Colucci, R., D'Erme, A.M., Moretti, S. & Lotti, T. Potential infectious etiology of Behçet's disease. Patholog. Res. Int. 2012, 595380 (2012).
Huuskonen, J., Olkkonen, V.M., Jauhiainen, M. & Ehnholm, C. The impact of phospholipid transfer protein (PLTP) on HDL metabolism. Atherosclerosis 155, 269–281 (2001).
Getz, G.S. & Reardon, C.A. Apoprotein E as a lipid transport and signaling protein in the blood, liver, and artery wall. J. Lipid Res. 50 (Suppl.), S156–S161 (2009).
Hevener, A.L. et al. Muscle-specific Pparg deletion causes insulin resistance. Nat. Med. 9, 1491–1497 (2003).
Hegele, R.A., Cao, H., Frankowski, C., Mathews, S.T. & Leff, T. PPARG F388L, a transactivation-deficient mutant, in familial partial lipodystrophy. Diabetes 51, 3586–3590 (2002).
Doney, A.S.F. et al. Association of the Pro12Ala and C1431T variants of PPARG and their haplotypes with susceptibility to type 2 diabetes. Diabetologia 47, 555–558 (2004).
Hao, H.-X. et al. ZNRF3 promotes Wnt receptor turnover in an R-spondin–sensitive manner. Nature 485, 195–200 (2012).
Farin, H.F. et al. Visualization of a short-range Wnt gradient in the intestinal stem-cell niche. Nature 530, 340–343 (2016).
Zhou, Y. et al. ZNRF3 acts as a tumour suppressor by the Wnt signalling pathway in human gastric adenocarcinoma. J. Mol. Histol. 44, 555–563 (2013).
Casals, F. et al. Genetic adaptation of the antibacterial human innate immunity network. BMC Evol. Biol. 11, 202 (2011).
Bunge, J., Willis, A. & Walsh, F. Estimating the number of species in microbial diversity studies. Annu. Rev. Stat. Appl. 1, 427–445 (2014).
Caballero, S. & Pamer, E.G. Microbiota-mediated inflammation and antimicrobial defense in the intestine. Annu. Rev. Immunol. 33, 227–256 (2015).
Singh, V. et al. Interplay between enterobactin, myeloperoxidase and lipocalin 2 regulates E. coli survival in the inflamed gut. Nat. Commun. 6, 7113 (2015).
Nguyen, H.T.T. et al. Crohn's disease–associated adherent invasive Escherichia coli modulate levels of microRNAs in intestinal epithelial cells to reduce autophagy. Gastroenterology 146, 508–519 (2014).
Sadaghian Sadabad, M. et al. The ATG16L1-T300A allele impairs clearance of pathosymbionts in the inflamed ileal mucosa of Crohn's disease patients. Gut 64, 1546–1552 (2014).
Dambuza, I.M. & Brown, G.D. C-type lectins in immunity: recent developments. Curr. Opin. Immunol. 32, 21–27 (2015).
Iliev, I. D. et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science 336, 1314–1317 (2012).
Enattah, N.S. et al. Evidence of still-ongoing convergence evolution of the lactase persistence T-13910 alleles in humans. Am. J. Hum. Genet. 81, 615–625 (2007).
Tishkoff, S.A. et al. Convergent adaptation of human lactase persistence in Africa and Europe. Nat. Genet. 39, 31–40 (2007).
Troelsen, J.T. Adult-type hypolactasia and regulation of lactase expression. Biochim. Biophys. Acta 1723, 19–32 (2005).
Streppel, M.T. et al. Relative validity of the food frequency questionnaire used to assess dietary intake in the Leiden Longevity Study. Nutr. J. 12, 75 (2013).
Siebelink, E., Geelen, A. & de Vries, J.H.M. Self-reported energy intake by FFQ compared with actual energy intake to maintain body weight in 516 adults. Br. J. Nutr. 106, 274–281 (2011).
Genome of the Netherlands Consortium. Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat. Genet. 46, 818–825 (2014).
Shah, T.S. et al. optiCall: a robust genotype-calling algorithm for rare, low-frequency and common variants. Bioinformatics 28, 1598–1603 (2012).
Auton, A. et al. A global reference for human genetic variation. Nature 526, 68–74 (2015).
Deelen, P. et al. Genotype harmonizer: automatic strand alignment and format conversion for genotype data integration. BMC Res. Notes 7, 901 (2014).
Delaneau, O., Zagury, J.-F. & Marchini, J. Improved whole-chromosome phasing for disease and population genetic studies. Nat. Methods 10, 5–6 (2013).
Howie, B., Marchini, J. & Stephens, M. Genotype imputation with thousands of genomes. G3 (Bethesda) 1, 457–470 (2011).
Deelen, P. et al. Improved imputation quality of low-frequency and rare variants in European samples using the 'Genome of The Netherlands'. Eur. J. Hum. Genet. 22, 1321–1326 (2014).
Jia, X. et al. Imputing amino acid polymorphisms in human leukocyte antigens. PLoS One 8, e64683 (2013).
Bolger, A.M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Truong, D.T. et al. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat. Methods 12, 902–903 (2015).
Dimmer, E.C. et al. The UniProt-GO Annotation database in 2011. Nucleic Acids Res. 40, D565–D570 (2012).
Gene Ontology Consortium. Gene Ontology Consortium: going forward. Nucleic Acids Res. 43, D1049–D1056 (2015).
Vatanen, T. et al. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell 165, 842–853 (2016).
Westra, H.-J. et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet. 45, 1238–1243 (2013).
Bonder, M.J., Luijk, R., Zhernakova, D.V. & Moed, M. Disease variants alter transcription factor levels and methylation of their binding sites. Preprint at bioRxiv 033084 (2015).
Robinson, M.J., Sancho, D., Slack, E.C., LeibundGut-Landmann, S. & Reis e Sousa, C. Myeloid C-type lectins in innate immunity. Nat. Immunol. 7, 1258–1265 (2006).
Reikine, S., Nguyen, J.B. & Modis, Y. Pattern recognition and signaling mechanisms of RIG-I and MDA5. Front. Immunol. 5, 342 (2014).
Whitlock, M.C. Combining probability from independent tests: the weighted Z-method is superior to Fisher's approach. J. Evol. Biol. 18, 1368–1373 (2005).
Ardlie, K.G. et al. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–660 (2015).
Zhernakova, D.V. et al. Hypothesis-free identification of modulators of genetic risk factors. Preprint at bioRxiv 033217 (2015).
Acknowledgements
We thank the participants and the staff of LifeLines-DEEP, 500FG and MIBS for their collaboration. We thank J. Dekens, M. Platteel, J. Pietersma and A. Maatman for management and technical support, and K. McIntyre and J. Senior for editing the manuscript.
This project was funded by grants from Top Institute Food and Nutrition, Wageningen, to C.W. (TiFN GH001), the Netherlands Organization for Scientific Research to J.F. (NWO-VIDI 864.13.013), L.F. (ZonMW-VIDI 917.14.374) and R.K.W. (ZonMW-VIDI 016.136.308), and CardioVasculair Onderzoek Nederland to M.H.H., M.G.N., A.Z. and J.F. (CVON 2012-03). A.Z. holds a Rosalind Franklin Fellowship (University of Groningen). This research received funding from the European Research Council under the European Union's Seventh Framework Programme: C.W. is supported by FP7/2007-2013/ERC Advanced Grant (agreement 2012-322698) and a Spinoza Prize from the Netherlands Organization for Scientific Research. M.G.N. holds an ERC Consolidator Grant (310372). L.F. has an FP7/2007-2013 grant (agreement 259867) and an ERC Starting Grant (637640, ImmRisk). Y.L. holds a Netherlands Organization for Scientific Research VENI grant (863.13.011).
Author information
Authors and Affiliations
Contributions
Conceptualization: A.Z., J.F., C.W. and M.J.B. Methodology: M.J.B., A.K., L.F., J.F., P.D., T.V. and M.S. Software: M.J.B., A.K., L.F., J.F., P.D., M.A.S. and D.V.Z. Formal analysis: M.J.B., A.K., J.F. and A.Z. Investigation: A.Z., J.F., M.J.B., A.K., F.I., D.V.Z., S.A.J., A.V.V., E.F.T., H.H. and M.C.C. Resources: C.W., A.Z., L.F., J.F., M.A.S., M.G.N., R.J.X., L.J. and A.A.M.M. Data curation: M.J.B., A.K., J.F., P.D., L.F., S.A.J. and Y.L. Writing–original draft: A.Z., J.F., M.J.B., A.K. and C.W. Writing–review and editing: M.J.B., A.K., E.F.T., Z.M., F.I., A.V.V., P.D., T.V., M.S., S.P.S., D.V.Z., S.A.J., M.J., M.O., M.A.S., M.C.C., Y.L., V.K., H.H., R.K.W., L.F., M.H.H., D.J., M.G.N., C.W., J.F. and A.Z. Visualization: A.K., M.J.B., A.Z. and J.F. Supervision: A.Z., J.F., C.W., L.F., R.K.W. and M.H.H. Project administration: A.Z., J.F., C.W., L.F., M.G.N., D.J., A.A.M.M. and S.P.S. Funding acquisition: A.Z., J.F., C.W., L.F., M.G.N., D.J. and A.A.M.M.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Genome-wide significant microbial QTL plots on the microbial level.
The plots show the effect of SNPs on normalized microbial abundance, showing a combination of violin plots and box plots. The box plots show the median and 25% and 75% quantiles.
Supplementary Figure 2 Genome-wide significant microbial QTL plots on microbial function level (MetaCyc-Pathway).
The plots show the effect of SNPs on normalized microbial functional abundance, showing a combination of violin plots and box plots. The box plots show the median and 25% and 75% quantiles.
Supplementary Figure 3 Genome-wide microbial QTL plots on the microbial function level (GO terms).
The plots show the effect of SNPs on normalized functional abundance, showing a combination of violin plots and box plots. The box plots show the median and 25% and 75% quantiles.
Supplementary Figure 4 Correlation of associated GO terms with taxonomies on the species level.
The plots show the Spearman correlations between the tested taxonomies and GO2000 terms.
Supplementary Figure 5 Relationship between Bifidobacterium and milk consumption and between an LCT SNP (rs4988235) and milk consumption.
(a) Correlation of milk consumption with Bifidobacterium abundance in the three tested cohorts. (b) Relationship of a functional lactase variant and milk consumption in the three cohorts.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5. (PDF 1520 kb)
Supplementary Table 1
Abundance levels of microbes, MetaCyc pathways and GO2000 terms. (XLSX 181 kb)
Supplementary Table 2
Summary of the tested number of associations per analysis branch. (XLSX 9 kb)
Supplementary Table 3
Genome-wide microbial QTL results. (XLSX 46 kb)
Supplementary Table 4
Estimations for the FDRs presented in the paper. (XLSX 12 kb)
Supplementary Table 5
Correlations between microbial abundance and MetaCyc abundance levels. (XLSX 1860 kb)
Supplementary Table 6
Correlations between microbial abundance and GO2000 abundance levels. (XLSX 1502 kb)
Supplementary Table 7
SNP selection list GWAS-associated SNPs. (XLSX 143 kb)
Supplementary Table 8
SNP selection list for innate immunity and food preference, including references. (XLSX 9 kb)
Supplementary Table 9
Microbial QTLs on abundance and functional levels for SNPs previously related to GWAS. (XLSX 17 kb)
Supplementary Table 10
Microbial QTLs on abundance and functional levels for the variants in the HLA. (XLSX 11 kb)
Supplementary Table 11
Microbial QTLs on abundance and functional levels for SNPs related to innate immunity. (XLSX 28 kb)
Supplementary Table 12
Microbial QTLs on abundance and functional levels for SNPs related to food preference. (XLSX 14 kb)
Rights and permissions
About this article
Cite this article
Bonder, M., Kurilshikov, A., Tigchelaar, E. et al. The effect of host genetics on the gut microbiome. Nat Genet 48, 1407–1412 (2016). https://doi.org/10.1038/ng.3663
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3663
This article is cited by
-
Human genetic associations of the airway microbiome in chronic obstructive pulmonary disease
Respiratory Research (2024)
-
Variant of the lactase LCT gene explains association between milk intake and incident type 2 diabetes
Nature Metabolism (2024)
-
A genome-wide association study reveals the relationship between human genetic variation and the nasal microbiome
Communications Biology (2024)
-
Scrutinizing microbiome determinism: why deterministic hypotheses about the microbiome are conceptually ungrounded
History and Philosophy of the Life Sciences (2024)
-
Analysis of strain, sex, and diet-dependent modulation of gut microbiota reveals candidate keystone organisms driving microbial diversity in response to American and ketogenic diets
Microbiome (2023)