Abstract
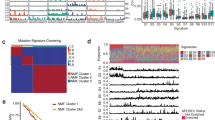
Esophageal adenocarcinoma (EAC) has a poor outcome, and targeted therapy trials have thus far been disappointing owing to a lack of robust stratification methods. Whole-genome sequencing (WGS) analysis of 129 cases demonstrated that this is a heterogeneous cancer dominated by copy number alterations with frequent large-scale rearrangements. Co-amplification of receptor tyrosine kinases (RTKs) and/or downstream mitogenic activation is almost ubiquitous; thus tailored combination RTK inhibitor (RTKi) therapy might be required, as we demonstrate in vitro. However, mutational signatures showed three distinct molecular subtypes with potential therapeutic relevance, which we verified in an independent cohort (n = 87): (i) enrichment for BRCA signature with prevalent defects in the homologous recombination pathway; (ii) dominant T>G mutational pattern associated with a high mutational load and neoantigen burden; and (iii) C>A/T mutational pattern with evidence of an aging imprint. These subtypes could be ascertained using a clinically applicable sequencing strategy (low coverage) as a basis for therapy selection.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Change history
19 September 2016
In the version of this article initially published online, the mutation signature illustrations for S1 and S2 in Figure 3a were switched. Additionally, in the Online Methods, the text originally stated that structural variants were called using BWA-MEM, when it should have stated that these were called using BWA. These errors have been corrected for the print, PDF and HTML versions of this article.
References
Ferlay, J. et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 136, E359–E386 (2015).
Brown, L.M., Devesa, S.S. & Chow, W.H. Incidence of adenocarcinoma of the esophagus among white Americans by sex, stage, and age. J. Natl. Cancer Inst. 100, 1184–1187 (2008).
Cunningham, D., Okines, A.F. & Ashley, S. Capecitabine and oxaliplatin for advanced esophagogastric cancer. N. Engl. J. Med. 362, 858–859 (2010).
Allum, W.H., Stenning, S.P., Bancewicz, J., Clark, P.I. & Langley, R.E. Long-term results of a randomized trial of surgery with or without preoperative chemotherapy in esophageal cancer. J. Clin. Oncol. 27, 5062–5067 (2009).
Cunningham, D. et al. Perioperative chemotherapy versus surgery alone for resectable gastroesophageal cancer. N. Engl. J. Med. 355, 11–20 (2006).
van Hagen, P. et al. Preoperative chemoradiotherapy for esophageal or junctional cancer. N. Engl. J. Med. 366, 2074–2084 (2012).
Bang, Y.J. et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet 376, 687–697 (2010).
Gao, Y.B. et al. Genetic landscape of esophageal squamous cell carcinoma. Nat. Genet. 46, 1097–1102 (2014).
Schulze, K. et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat. Genet. 47, 505–511 (2015).
Cancer Genome Atlas Network. Genomic classification of cutaneous melanoma. Cell 161, 1681–1696 (2015).
Waddell, N. et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 518, 495–501 (2015).
Totoki, Y. et al. Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nat. Genet. 46, 1267–1273 (2014).
Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 513, 202–209 (2014).
Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550 (2014).
Chantrill, L.A. et al. Precision medicine for advanced pancreas cancer: The Individualized Molecular Pancreatic Cancer Therapy (IMPaCT) Trial. Clin. Cancer Res. 21, 2029–2037 (2015).
Dulak, A.M. et al. Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nat. Genet. 45, 478–486 (2013).
Weaver, J.M. et al. Ordering of mutations in preinvasive disease stages of esophageal carcinogenesis. Nat. Genet. 46, 837–843 (2014).
Nones, K. et al. Genomic catastrophes frequently arise in esophageal adenocarcinoma and drive tumorigenesis. Nat. Commun. 5, 5224 (2014).
Cancer Genome Atlas Research Network. et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 45, 1113–1120 (2013).
Paterson, A.L. et al. Mobile element insertions are frequent in oesophageal adenocarcinomas and can mislead paired-end sequencing analysis. BMC Genomics 16, 473 (2015).
Tubio, J.M. et al. Mobile DNA in cancer. Extensive transduction of nonrepetitive DNA mediated by L1 retrotransposition in cancer genomes. Science 345, 1251343 (2014).
Mermel, C.H. et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 12, R41 (2011).
Lawrence, M.S. et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218 (2013).
Nik-Zainal, S. et al. The life history of 21 breast cancers. Cell 149, 994–1007 (2012).
Paterson, A.L. et al. Characterization of the timing and prevalence of receptor tyrosine kinase expression changes in oesophageal carcinogenesis. J. Pathol. 230, 118–128 (2013).
Van Cutsem, E. et al. HER2 screening data from ToGA: targeting HER2 in gastric and gastroesophageal junction cancer. Gastric Cancer 18, 476–484 (2015).
Alexandrov, L.B. et al. Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013).
Shiraishi, Y., Tremmel, G., Miyano, S. & Stephens, M. A simple model-based approach to inferring and visualizing cancer mutation signatures. PLoS Genet. 11, e1005657 (2015).
Gehring, J.S., Fischer, B., Lawrence, M. & Huber, W. SomaticSignatures: inferring mutational signatures from single-nucleotide variants. Bioinformatics 31, 3673–3675 (2015).
Pearl, L.H., Schierz, A.C., Ward, S.E., Al-Lazikani, B. & Pearl, F.M. Therapeutic opportunities within the DNA damage response. Nat. Rev. Cancer 15, 166–180 (2015).
Shen, J. et al. ARID1A deficiency impairs the DNA damage checkpoint and sensitizes cells to PARP inhibitors. Cancer Discov. 5, 752–767 (2015).
Hodi, F.S. et al. Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med. 363, 711–723 (2010).
Larkin, J. et al. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N. Engl. J. Med. 373, 23–34 (2015).
Herbst, R.S. et al. Pembrolizumab versus docetaxel for previously treated, PD-L1–positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet 387, 1540–1550 (2016).
Rizvi, N.A. et al. Mutational landscape determines sensitivity to PD-1 blockade in non–small cell lung cancer. Science 348, 124–128 (2015).
Snyder, A. et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N. Engl. J. Med. 371, 2189–2199 (2014).
McGranahan, N. et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 351, 1463–1469 (2016).
Van Allen, E.M. et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 350, 207–211 (2015).
Tumeh, P.C. et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 515, 568–571 (2014).
Hamanishi, J. et al. Programmed cell death 1 ligand 1 and tumor-infiltrating CD8+ T lymphocytes are prognostic factors of human ovarian cancer. Proc. Natl. Acad. Sci. USA 104, 3360–3365 (2007).
Benafif, S. & Hall, M. An update on PARP inhibitors for the treatment of cancer. Onco Targets Ther. 8, 519–528 (2015).
Oza, A.M. et al. Olaparib combined with chemotherapy for recurrent platinum-sensitive ovarian cancer: a randomised phase 2 trial. Lancet Oncol. 16, 87–97 (2015).
Demel, H.R. et al. Effects of topoisomerase inhibitors that induce DNA damage response on glucose metabolism and PI3K/Akt/mTOR signaling in multiple myeloma cells. Am. J. Cancer Res. 5, 1649–1664 (2015).
Farmer, H. et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434, 917–921 (2005).
Di Leonardo, A., Linke, S.P., Clarkin, K. & Wahl, G.M. DNA damage triggers a prolonged p53-dependent G1 arrest and long-term induction of Cip1 in normal human fibroblasts. Genes Dev. 8, 2540–2551 (1994).
Agarwal, M.L. et al. A p53-dependent S-phase checkpoint helps to protect cells from DNA damage in response to starvation for pyrimidine nucleotides. Proc. Natl. Acad. Sci. USA 95, 14775–14780 (1998).
Brooks, K. et al. A potent Chk1 inhibitor is selectively cytotoxic in melanomas with high levels of replicative stress. Oncogene 32, 788–796 (2013).
Vera, J. et al. Chk1 and Wee1 control genotoxic-stress induced G2-M arrest in melanoma cells. Cell. Signal. 27, 951–960 (2015).
Liu, Q. et al. Chk1 is an essential kinase that is regulated by Atr and required for the G2/M DNA damage checkpoint. Genes Dev. 14, 1448–1459 (2000).
Watanabe, N., Broome, M. & Hunter, T. Regulation of the human WEE1Hu CDK tyrosine 15-kinase during the cell cycle. EMBO J. 14, 1878–1891 (1995).
van de Wetering, M. et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 161, 933–945 (2015).
Sato, T. et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 (2009).
Ciriello, G. et al. Emerging landscape of oncogenic signatures across human cancers. Nat. Genet. 45, 1127–1133 (2013).
Osato, M. Point mutations in the RUNX1/AML1 gene: another actor in RUNX leukemia. Oncogene 23, 4284–4296 (2004).
Watkins, J.A., Irshad, S., Grigoriadis, A. & Tutt, A.N. Genomic scars as biomarkers of homologous recombination deficiency and drug response in breast and ovarian cancers. Breast Cancer Res. 16, 211 (2014).
Alexandrov, L.B., Nik-Zainal, S., Siu, H.C., Leung, S.Y. & Stratton, M.R. A mutational signature in gastric cancer suggests therapeutic strategies. Nat. Commun. 6, 8683 (2015).
Ledermann, J. et al. Olaparib maintenance therapy in patients with platinum-sensitive relapsed serous ovarian cancer: a preplanned retrospective analysis of outcomes by BRCA status in a randomised phase 2 trial. Lancet Oncol. 15, 852–861 (2014).
Verhagen, C.V. et al. Extent of radiosensitization by the PARP inhibitor olaparib depends on its dose, the radiation dose and the integrity of the homologous recombination pathway of tumor cells. Radiother. Oncol. 116, 358–365 (2015).
Kelly, R.J. et al. Adaptive immune resistance in gastro-esophageal cancer: correlating tumoral/stromal PDL1 expression with CD8+ cell count. J. Clin. Oncol. 33, abstr. 4031 (2015).
Nakamura, H. et al. Genomic spectra of biliary tract cancer. Nat. Genet. 47, 1003–1010 (2015).
Bridges, K.A. et al. MK-1775, a novel Wee1 kinase inhibitor, radiosensitizes p53-defective human tumor cells. Clin. Cancer Res. 17, 5638–5648 (2011).
Wang, Y. et al. Radiosensitization of p53 mutant cells by PD0166285, a novel G2 checkpoint abrogator. Cancer Res. 61, 8211–8217 (2001).
Liu, D.S. et al. APR-246 potently inhibits tumour growth and overcomes chemoresistance in preclinical models of oesophageal adenocarcinoma. Gut 64, 1506–1516 (2015).
Stewart, A., Thavasu, P., de Bono, J.S. & Banerji, U. Titration of signalling output: insights into clinical combinations of MEK and AKT inhibitors. Ann. Oncol. 26, 1504–1510 (2015).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Saunders, C.T. et al. Strelka: accurate somatic small-variant calling from sequenced tumor–normal sample pairs. Bioinformatics 28, 1811–1817 (2012).
McLaren, W. et al. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics 26, 2069–2070 (2010).
Van Loo, P. et al. Allele-specific copy number analysis of tumors. Proc. Natl. Acad. Sci. USA 107, 16910–16915 (2010).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Zack, T.I. et al. Pan-cancer patterns of somatic copy number alteration. Nat. Genet. 45, 1134–1140 (2013).
Boeva, V. et al. Control-FREEC: a tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics 28, 423–425 (2012).
Chen, X. et al. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics 32, 1220–1222 (2016).
Schulte, I. et al. Structural analysis of the genome of breast cancer cell line ZR-75-30 identifies twelve expressed fusion genes. BMC Genomics 13, 719 (2012).
Le Tallec, B. et al. Common fragile site profiling in epithelial and erythroid cells reveals that most recurrent cancer deletions lie in fragile sites hosting large genes. Cell Reports 4, 420–428 (2013).
Auton, A. et al. A global reference for human genetic variation. Nature 526, 68–74 (2015).
Wilkerson, M.D. & Hayes, D.N. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics 26, 1572–1573 (2010).
Nilsen, G. et al. Copynumber: efficient algorithms for single- and multi-track copy number segmentation. BMC Genomics 13, 591 (2012).
Korbel, J.O. & Campbell, P.J. Criteria for inference of chromothripsis in cancer genomes. Cell 152, 1226–1236 (2013).
Puente, X.S. et al. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature 526, 519–524 (2015).
Kumar, P., Henikoff, S. & Ng, P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 4, 1073–1081 (2009).
Adzhubei, I.A. et al. A method and server for predicting damaging missense mutations. Nat. Methods 7, 248–249 (2010).
Lundegaard, C. et al. NetMHC-3.0: accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8–11. Nucleic Acids Res. 36, W509–W512 (2008).
Adiconis, X. et al. Comparative analysis of RNA sequencing methods for degraded or low-input samples. Nat. Methods 10, 623–629 (2013).
Acknowledgements
This paper is dedicated to Nadeera de Silva, who tragically and unexpectedly died while this paper was undergoing revision. He made an important contribution to this research, particularly bringing his clinical oncology perspective to bear on the translational relevance of the findings.
Whole-genome sequencing of esophageal adenocarcinoma samples was carried out in concert with the International Cancer Genome Consortium (ICGC) through the OCCAMS Consortium and was funded by program grants from Cancer Research UK (RG66287, RG81771, RG84119). We thank the ICGC members for their input on verification standards as part of the benchmarking exercise. We thank the Human Research Tissue Bank, which is supported by the National Institute for Health Research (NIHR) Cambridge Biomedical Research Centre, from Addenbrooke's Hospital and UCL. We also thank the University Hospital of Southampton Trust; the Southampton, Birmingham, Edinburgh and UCL Experimental Cancer Medicine Centres; and the QEHB charities. R.C.F. is funded by an NIHR Professorship (RG67258) and receives core funding from the Medical Research Council (RG84369) and infrastructure support from the Biomedical Research Centre (RG64237) and the Experimental Cancer Medicine Centre (RG62923). We acknowledge the support of the University of Cambridge, Cancer Research UK (C14303/A17197) and Hutchison Whampoa Limited. We thank P. Van Loo for providing the NGS version of ASCAT for copy number calling. We are grateful to all the patients who provided written consent for participation in this study and to the staff at all participating centres.
Some of the work was undertaken at UCLH/UCL, which received a proportion of funding from the Department of Health's NIHR Biomedical Research Centres funding scheme. The views expressed in this publication are those of the authors and are not necessarily those of the Department of Health. The work at UCLH/UCL was also supported by the CRUK UCL Early Cancer Medicine Centre.
Author information
Authors and Affiliations
Consortia
Contributions
R.C.F. conceived the overall study. M.S., X.L. and P.A.W.E. analyzed the data. R.C.F., M.S., X.L., N.d.S., P.A.W.E. and A.G.L. conceived and designed the experiments. M.S. performed the statistical analysis. X.L., G.C., S.M., M.O., A.M., J.C. and N.G.-D. performed the experiments. M.D.E. performed benchmarking studies on the variant calls, and implemented and ran several variant-calling and analysis pipelines. G.C. contributed to the structural variant analysis. J.B. contributed expression data and curated the clinical data collection. S.M. and N.G. coordinated sample processing with clinical centers and was responsible for sample collections. T.-P.Y. performed the BFB analysis. L.B. ran the variant-calling pipelines. H.C. contributed to the RTK analysis. A.G., J.S. and T.U. contributed cell lines. N.W. and A.P.B. contributed sequencing data for validation. B.N. coordinated data and tissue collection from centers for the study. A.A. helped develop the copy-number-calling pipeline. R.C.F. and S.T. jointly supervised the research. M.S., N.d.S., X.L. and R.C.F. wrote the manuscript. All authors approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–28, Supplementary Tables 1, 2, 4–9, 11 and 12 and Supplementary Note (PDF 5505 kb)
Supplementary Table 3
Significantly deleted loci in the cohort according to GISTIC2.0. Loci with residual q-value (XLSX 13 kb)
Supplementary Table 10
Microsatellite instability analysis results. The potentially microsatellite unstable samples that were removed from the analysis are highlighted at the top. (XLSX 13 kb)
Rights and permissions
About this article
Cite this article
Secrier, M., Li, X., de Silva, N. et al. Mutational signatures in esophageal adenocarcinoma define etiologically distinct subgroups with therapeutic relevance. Nat Genet 48, 1131–1141 (2016). https://doi.org/10.1038/ng.3659
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3659
This article is cited by
-
A decade of the Oesophageal Cancer Clinical and Molecular Stratification Consortium
Nature Medicine (2024)
-
Current status of vaccine immunotherapy for gastrointestinal cancers
Surgery Today (2023)
-
Multi-omic features of oesophageal adenocarcinoma in patients treated with preoperative neoadjuvant therapy
Nature Communications (2023)
-
Mutational signature dynamics shaping the evolution of oesophageal adenocarcinoma
Nature Communications (2023)
-
Comprehensive RNA dataset of tissue and plasma from patients with esophageal cancer or precursor lesions
Scientific Data (2022)