Abstract
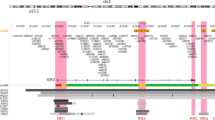
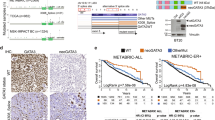
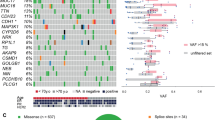
Sustained expression of the estrogen receptor-α (ESR1) drives two-thirds of breast cancer and defines the ESR1-positive subtype. ESR1 engages enhancers upon estrogen stimulation to establish an oncogenic expression program1. Somatic copy number alterations involving the ESR1 gene occur in approximately 1% of ESR1-positive breast cancers2,3,4,5, suggesting that other mechanisms underlie the persistent expression of ESR1. We report significant enrichment of somatic mutations within the set of regulatory elements (SRE) regulating ESR1 in 7% of ESR1-positive breast cancers. These mutations regulate ESR1 expression by modulating transcription factor binding to the DNA. The SRE includes a recurrently mutated enhancer whose activity is also affected by rs9383590, a functional inherited single-nucleotide variant (SNV) that accounts for several breast cancer risk–associated loci. Our work highlights the importance of considering the combinatorial activity of regulatory elements as a single unit to delineate the impact of noncoding genetic alterations on single genes in cancer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Primary accessions
Gene Expression Omnibus
Referenced accessions
Gene Expression Omnibus
References
Green, K.A. & Carroll, J.S. Oestrogen-receptor-mediated transcription and the influence of co-factors and chromatin state. Nat. Rev. Cancer 7, 713–722 (2007).
Vincent-Salomon, A., Raynal, V., Lucchesi, C., Gruel, N. & Delattre, O. ESR1 gene amplification in breast cancer: a common phenomenon? Nat. Genet. 40, 809, author reply 810–812 (2008).
Brown, L.A. et al. ESR1 gene amplification in breast cancer: a common phenomenon? Nat. Genet. 40, 806–807, author reply 810–812 (2008).
Horlings, H.M. et al. ESR1 gene amplification in breast cancer: a common phenomenon? Nat. Genet. 40, 807–808, author reply 810–812 (2008).
Reis-Filho, J.S. et al. ESR1 gene amplification in breast cancer: a common phenomenon? Nat. Genet. 40, 809–810, author reply 810–812 (2008).
Cowper-Sallari, R. et al. Breast cancer risk-associated SNPs modulate the affinity of chromatin for FOXA1 and alter gene expression. Nat. Genet. 44, 1191–1198 (2012).
Maurano, M.T. et al. Systematic localization of common disease-associated variation in regulatory DNA. Science 337, 1190–1195 (2012).
Schaub, M.A., Boyle, A.P., Kundaje, A., Batzoglou, S. & Snyder, M. Linking disease associations with regulatory information in the human genome. Genome Res. 22, 1748–1759 (2012).
Polak, P. et al. Reduced local mutation density in regulatory DNA of cancer genomes is linked to DNA repair. Nat. Biotechnol. 32, 71–75 (2014).
Polak, P. et al. Cell-of-origin chromatin organization shapes the mutational landscape of cancer. Nature 518, 360–364 (2015).
Weedon, M.N. et al. Recessive mutations in a distal PTF1A enhancer cause isolated pancreatic agenesis. Nat. Genet. 46, 61–64 (2014).
Horn, S. et al. TERT promoter mutations in familial and sporadic melanoma. Science 339, 959–961 (2013).
Huang, F.W. et al. Highly recurrent TERT promoter mutations in human melanoma. Science 339, 957–959 (2013).
Zheng, W. et al. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat. Genet. 41, 324–328 (2009).
Fletcher, O. et al. Novel breast cancer susceptibility locus at 9q31.2: results of a genome-wide association study. J. Natl. Cancer Inst. 103, 425–435 (2011).
Turnbull, C. et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat. Genet. 42, 504–507 (2010).
Siddiq, A. et al. A meta-analysis of genome-wide association studies of breast cancer identifies two novel susceptibility loci at 6q14 and 20q11. Hum. Mol. Genet. 21, 5373–5384 (2012).
Stacey, S.N. et al. Ancestry-shift refinement mapping of the C6orf97–ESR1 breast cancer susceptibility locus. PLoS Genet. 6, e1001029 (2010).
ENCODE Project Consortium. A user's guide to the Encyclopedia of DNA Elements (ENCODE). PLoS Biol. 9, e1001046 (2011).
1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073 (2010).
Thurman, R.E. et al. The accessible chromatin landscape of the human genome. Nature 489, 75–82 (2012).
Bailey, S.D. et al. ZNF143 provides sequence specificity to secure chromatin interactions at gene promoters. Nat. Commun. 2, 6186 (2015).
Curtis, C. et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486, 346–352 (2012).
Li, Q. et al. Integrative eQTL-based analyses reveal the biology of breast cancer risk loci. Cell 152, 633–641 (2013).
Dunning, A.M. et al. Breast cancer risk variants at 6q25 display different phenotype associations and regulate ESR1, RMND1 and CCDC170. Nat. Genet. 48, 374–386 (2016).
Frietze, S. et al. Cell type–specific binding patterns reveal that TCF7L2 can be tethered to the genome by association with GATA3. Genome Biol. 13, R52 (2012).
Alexandrov, L.B. et al. Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013).
Sanyal, A., Lajoie, B.R., Jain, G. & Dekker, J. The long-range interaction landscape of gene promoters. Nature 489, 109–113 (2012).
Ernst, J. & Kellis, M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat. Biotechnol. 28, 817–825 (2010).
Brocchieri, L. & Karlin, S. Protein length in eukaryotic and prokaryotic proteomes. Nucleic Acids Res. 33, 3390–3400 (2005).
Bell, R.J. et al. The transcription factor GABP selectively binds and activates the mutant TERT promoter in cancer. Science 348, 1036–1039 (2015).
Dunbier, A.K. et al. ESR1 is co-expressed with closely adjacent uncharacterised genes spanning a breast cancer susceptibility locus at 6q25.1. PLoS Genet. 7, e1001382 (2011).
Yamamoto-Ibusuki, M. et al. C6ORF97–ESR1 breast cancer susceptibility locus: influence on progression and survival in breast cancer patients. Eur. J. Hum. Genet. 23, 949–956 (2015).
Vogelstein, B. et al. Cancer genome landscapes. Science 339, 1546–1558 (2013).
Katainen, R. et al. CTCF/cohesin-binding sites are frequently mutated in cancer. Nat. Genet. 47, 818–821 (2015).
Taberlay, P.C., Statham, A.L., Kelly, T.K., Clark, S.J. & Jones, P.A. Reconfiguration of nucleosome-depleted regions at distal regulatory elements accompanies DNA methylation of enhancers and insulators in cancer. Genome Res. 24, 1421–1432 (2014).
Korn, J.M. et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat. Genet. 40, 1253–1260 (2008).
Danecek, P. et al. The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Trapnell, C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012).
Bolstad, B.M., Irizarry, R.A., Astrand, M. & Speed, T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19, 185–193 (2003).
Bailey, S.D., Virtanen, C., Haibe-Kains, B. & Lupien, M. ABC: a tool to identify SNVs causing allele-specific transcription factor binding from ChIP-Seq experiments. Bioinformatics 31, 3057–3059 (2015).
Quinlan, A.R. & Hall, I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
John, S. et al. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat. Genet. 43, 264–268 (2011).
Lawrence, M.S. et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218 (2013).
Dees, N.D. et al. MuSiC: identifying mutational significance in cancer genomes. Genome Res. 22, 1589–1598 (2012).
Stergachis, A.B. et al. Exonic transcription factor binding directs codon choice and affects protein evolution. Science 342, 1367–1372 (2013).
Ju, Y.S. et al. Frequent somatic transfer of mitochondrial DNA into the nuclear genome of human cancer cells. Genome Res. 25, 814–824 (2015).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Cibulskis, K. et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 31, 213–219 (2013).
Kulakovskiy, I.V. et al. HOCOMOCO: a comprehensive collection of human transcription factor binding sites models. Nucleic Acids Res. 41, D195–D202 (2013).
Grant, C.E., Bailey, T.L. & Noble, W.S. FIMO: scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018 (2011).
Livak, K.J. & Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔC T) method. Methods 25, 402–408 (2001).
Li, B., Kadura, I., Fu, D.J. & Watson, D.E. Genotyping with TaqMAMA. Genomics 83, 311–320 (2004).
Hagège, H. et al. Quantitative analysis of chromosome conformation capture assays (3C-qPCR). Nat. Protoc. 2, 1722–1733 (2007).
Acknowledgements
We thank A. Razak, C. Elser, D. Cescon, D. Warr, E. Amir, L. Siu, N. Leighl and S. Sridhar for their involvement in recruiting the IMPACT and COMPACT samples used in this study. We also thank M. Lemaire for helpful discussions. We thank R. Rottapel and O. Kent for use of and help with the Glomax Multi-Detection system. We acknowledge the ENCODE consortium and the ENCODE production laboratories that generated the data sets provided by the ENCODE Data Coordination Center used in the manuscript. We also acknowledge the Cancer Genome Project, for making all the breast cancer and liver cancer called mutations publicly available, and the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC), for making the genotyping and expression data from primary breast tumors data available. We acknowledge the Princess Margaret Genomics Centre and the Bioinformatics group for providing the infrastructure assisting us with the targeted sequencing and analysis of the ESR1 SRE. Supported by the National Cancer Institute (NCI) at the National Institute of Health (NIH) (R01CA155004 to M.L.), the Princess Margaret Cancer Foundation (T.J.P. and M.L.), The Canadian Cancer Society (CCSRI702922 to M.L.), the Susan G. Komen Foundation (CCR15332792 to T.J.P.) and the Gattuso-Slaight Personalized Cancer Medicine Fund/PMCF (B.H.-K.). M.L. is funded by a young investigator award from the Ontario Institute for Cancer Research (OICR), a new investigator salary award from the Canadian Institute of Health Research (CIHR) and a Movember Rising Star award from Prostate Cancer Canada (PCC) (RS2014-04). K.J.K. and R.C.P. are supported by Canadian Breast Cancer Foundation (CBCF) postdoctoral fellowships. S.D.B. is supported by a Knudson and CIHR postdoctoral fellowship.
Author information
Authors and Affiliations
Contributions
The concept of interrogating the mutational load in regulatory elements converging on single genes arose through discussions between S.D.B., N.A.S.-A., R.C.S. and M.L. S.D.B. designed and/or implemented all the computational and statistical approaches except for IGR and analyzed the results under the supervision of M.L. Experimental assessment of the effect of SNVs on enhancer activity, transcription factor binding and gene expression was designed by K.D., S.D.B. and M.L. and conducted by K.D. with assistance from K.J.K., A.E.T. and X.W. The CRISPR–Cas9-based enhancer deletion was conducted by K.D., K.J.K., K.L.T., J.S. and D.W.C. under the supervision of T.W.M. and M.L. P.M. and N.A.S.-A. implemented the IGR approach to predict allele-bias binding of transcription factors on SNVs after improvements to IGR by N.A.S.-A. and R.C.S. R.C.P. and P.L.B. assessed the ESR1, PR and HER2 expression status on primary breast tumors included in our validation cohort. S.Y.C.Y. performed the alignment and gene expression quantification of the TCGA RNA-seq data. M.D. assisted in DNA capture sequencing of the primary breast tumor validation cohort under T.J.P.'s supervision. B.H.-K. oversaw the expression analysis of the METABRIC data set. M.L. oversaw the project. Figures were designed and prepared by S.D.B. and K.D. The manuscript was written by S.D.B., K.D. and M.L. with assistance from all other authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–17, Supplementary Tables 1–9 (PDF 9695 kb)
Rights and permissions
About this article
Cite this article
Bailey, S., Desai, K., Kron, K. et al. Noncoding somatic and inherited single-nucleotide variants converge to promote ESR1 expression in breast cancer. Nat Genet 48, 1260–1266 (2016). https://doi.org/10.1038/ng.3650
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3650
This article is cited by
-
Non-coding driver mutations in human cancer
Nature Reviews Cancer (2021)
-
Intrinsic and Extrinsic Factors Governing the Transcriptional Regulation of ESR1
Hormones and Cancer (2020)
-
Noncoding mutations target cis-regulatory elements of the FOXA1 plexus in prostate cancer
Nature Communications (2020)
-
Japanese GWAS identifies variants for bust-size, dysmenorrhea, and menstrual fever that are eQTLs for relevant protein-coding or long non-coding RNAs
Scientific Reports (2018)
-
CRISPR-based strategies for studying regulatory elements and chromatin structure in mammalian gene control
Mammalian Genome (2018)