Abstract
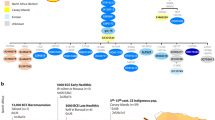
To shed light on the peopling of South Asia and the origins of the morphological adaptations found there, we analyzed whole-genome sequences from 10 Andamanese individuals and compared them with sequences for 60 individuals from mainland Indian populations with different ethnic histories and with publicly available data from other populations. We show that all Asian and Pacific populations share a single origin and expansion out of Africa, contradicting an earlier proposal of two independent waves of migration1,2,3,4. We also show that populations from South and Southeast Asia harbor a small proportion of ancestry from an unknown extinct hominin, and this ancestry is absent from Europeans and East Asians. The footprints of adaptive selection in the genomes of the Andamanese show that the characteristic distinctive phenotypes of this population (including very short stature) do not reflect an ancient African origin but instead result from strong natural selection on genes related to human body size.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
References
Coon, C.S. & Hunt, E.E. The Living Races of Man (Knopf, 1966).
Molnar, S. Human Variation: Races, Types and Ethnic Groups (Routledge, 2015).
Cavalli-Sforza, L.L., Menozzi, P. & Piazza, A. The History and Geography of Human Genes (Princeton University Press, 1994).
Rasmussen, M. et al. An Aboriginal Australian genome reveals separate human dispersals into Asia. Science 334, 94–98 (2011).
Huxley, T.H. On the geographical distribution of the chief modifications of mankind. J. Ethnol. Soc. London 2, 404–412 (1870).
Brown, A.R. The Andaman Islanders: A Study in Social Anthropology (Cambridge University Press, 1922).
Abbi, A. Is Great Andamanese genealogically and typologically distinct from Onge and Jarawa? Lang. Sci. 31, 791–812 (2009).
Reich, D., Thangaraj, K., Patterson, N., Price, A.L. & Singh, L. Reconstructing Indian population history. Nature 461, 489–494 (2009).
Montgomery, S.H. Primate brains, the 'island rule' and the evolution of Homo floresiensis. J. Hum. Evol. 65, 750–760 (2013).
Zoledziewska, M. et al. Height-reducing variants and selection for short stature in Sardinia. Nat. Genet. 47, 1352–1356 (2015).
Basu, A., Sarkar-Roy, N. & Majumder, P.P. Genomic reconstruction of the history of extant populations of India reveals five distinct ancestral components and a complex structure. Proc. Natl. Acad. Sci. USA 113, 1594–1599 (2016).
Dass, F. The Andaman Islands (The Good Shepherd Convent Press, 1937).
Green, R.E. et al. A draft sequence of the Neandertal genome. Science 328, 710–722 (2010).
Pickrell, J.K. & Pritchard, J.K. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 8, e1002967 (2012).
Schiffels, S. & Durbin, R. Inferring human population size and separation history from multiple genome sequences. Nat. Genet. 46, 919–925 (2014).
Raghavan, M. et al. Upper Palaeolithic Siberian genome reveals dual ancestry of Native Americans. Nature 505, 87–91 (2014).
Olalde, I. et al. Derived immune and ancestral pigmentation alleles in a 7,000-year-old Mesolithic European. Nature 507, 225–228 (2014).
Lazaridis, I. et al. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature 513, 409–413 (2014).
Haak, W. et al. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature 522, 207–211 (2015).
Prüfer, K. et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505, 43–49 (2014).
Meyer, M. et al. A high-coverage genome sequence from an archaic Denisovan individual. Science 338, 222–226 (2012).
Swisher, C.C. III et al. Latest Homo erectus of Java: potential contemporaneity with Homo sapiens in southeast Asia. Science 274, 1870–1874 (1996).
Hammer, M.F., Woerner, A.E., Mendez, F.L., Watkins, J.C. & Wall, J.D. Genetic evidence for archaic admixture in Africa. Proc. Natl. Acad. Sci. USA 108, 15123–15128 (2011).
Vernot, B. & Akey, J.M. Resurrecting surviving Neandertal lineages from modern human genomes. Science 343, 1017–1021 (2014).
Juyal, G. et al. Population and genomic lessons from genetic analysis of two Indian populations. Hum. Genet. 133, 1273–1287 (2014).
Pybus, M. et al. Hierarchical boosting: a machine-learning framework to detect and classify hard selective sweeps in human populations. Bioinformatics 31, 3946–3952 (2015).
Becker, K.G., Barnes, K.C., Bright, T.J. & Wang, S.A. The genetic association database. Nat. Genet. 36, 431–432 (2004).
1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 (2012).
Prado-Martinez, J. et al. Great ape genetic diversity and population history. Nature 499, 471–475 (2013).
Sudmant, P.H. et al. Global diversity, population stratification, and selection of human copy-number variation. Science 349, aab3761 (2015).
Drmanac, R. et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science 327, 78–81 (2010).
1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073 (2010).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Manichaikul, A. et al. Robust relationship inference in genome-wide association studies. Bioinformatics 26, 2867–2873 (2010).
Danecek, P. et al. The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Price, A.L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006).
Alexander, D.H., Novembre, J. & Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664 (2009).
Delaneau, O., Marchini, J. & 1000 Genomes Project Consortium Integrating sequence and array data to create an improved 1000 Genomes Project haplotype reference panel. Nat. Commun. 5, 3934 (2014).
Patterson, N. et al. Ancient admixture in human history. Genetics 192, 1065–1093 (2012).
Hudson, R.R. Generating samples under a Wright–Fisher neutral model of genetic variation. Bioinformatics 18, 337–338 (2002).
Gravel, S. et al. Demographic history and rare allele sharing among human populations. Proc. Natl. Acad. Sci. USA 108, 11983–11988 (2011).
Gutenkunst, R.N., Hernandez, R.D., Williamson, S.H. & Bustamante, C.D. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 5, e1000695 (2009).
Delaneau, O., Marchini, J. & Zagury, J.-F. Alinear complexity phasing method for thousands of genomes. Nat. Methods 9, 179–181 (2012).
Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595 (1989).
Nielsen, R. et al. Genomic scans for selective sweeps using SNP data. Genome Res. 15, 1566–1575 (2005).
Fay, J.C. & Wu, C.I. Hitchhiking under positive Darwinian selection. Genetics 155, 1405–1413 (2000).
Fu, Y.X. & Li, W.H. Statistical tests of neutrality of mutations. Genetics 133, 693–709 (1993).
Sabeti, P.C. et al. Genome-wide detection and characterization of positive selection in human populations. Nature 449, 913–918 (2007).
Voight, B.F., Kudaravalli, S., Wen, X. & Pritchard, J.K. A map of recent positive selection in the human genome. PLoS Biol. 4, e72 (2006).
Sabeti, P.C. et al. Detecting recent positive selection in the human genome from haplotype structure. Nature 419, 832–837 (2002).
Schaffner, S.F. et al. Calibrating a coalescent simulation of human genome sequence variation. Genome Res. 15, 1576–1583 (2005).
Acknowledgements
J. Nye and C. Tyler-Smith kindly corrected the manuscript in depth. Thanks are given to R.A. Foley for discussion and inspiring input for Figure 3. Our main funding was provided by the joint Spain–India bilateral grant PRI-PIBIN-2011-0942 from the Ministerio de Economía y Competitividad (Spain). Complementary funding was provided by grant BFU2013-43726-P from the Ministerio de Economía y Competitividad (Spain), with the support of Secretaria d'Universitats i Recerca, Departament d'Economia i Coneixement de la Generalitat de Catalunya (GRC 2014 SGR866).
Author information
Authors and Affiliations
Contributions
M.M., F.C., P.P.M. and J.B. conceived and designed the project. P.P.M. provided the samples. P.P.M., T.X. and Q.L. sequenced samples and carried out initial analyses. M.M. performed the remaining genetic data analyses. F.C., G.M.D., M.P., M.G.N., D.C., H.L., P.P.M. and J.B. participated in and discussed analyses. M.M., F.C., P.P.M. and J.B. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–38, Supplementary Tables 1–17 and Supplementary Note. (PDF 6962 kb)
Rights and permissions
About this article
Cite this article
Mondal, M., Casals, F., Xu, T. et al. Genomic analysis of Andamanese provides insights into ancient human migration into Asia and adaptation. Nat Genet 48, 1066–1070 (2016). https://doi.org/10.1038/ng.3621
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3621
This article is cited by
-
The Allen Ancient DNA Resource (AADR) a curated compendium of ancient human genomes
Scientific Data (2024)
-
Population history modulates the fitness effects of Copy Number Variation in the Roma
Human Genetics (2023)
-
The coefficients of inbreeding revealed by ROH study among inbred individuals belonging to each type of the first cousin marriage: A preliminary report from North India
Genes & Genomics (2023)
-
Early presence of Homo sapiens in Southeast Asia by 86–68 kyr at Tam Pà Ling, Northern Laos
Nature Communications (2023)
-
Counteracting forces of introgressive hybridization and interspecific competition shape the morphological traits of cryptic Iberian Eptesicus bats
Scientific Reports (2022)