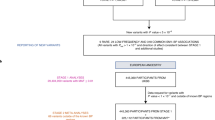
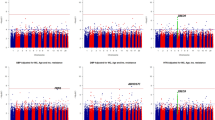
Abstract
Elevated blood pressure is a common, heritable cause of cardiovascular disease worldwide. To date, identification of common genetic variants influencing blood pressure has proven challenging. We tested 2.5 million genotyped and imputed SNPs for association with systolic and diastolic blood pressure in 34,433 subjects of European ancestry from the Global BPgen consortium and followed up findings with direct genotyping (N ≤ 71,225 European ancestry, N ≤ 12,889 Indian Asian ancestry) and in silico comparison (CHARGE consortium, N = 29,136). We identified association between systolic or diastolic blood pressure and common variants in eight regions near the CYP17A1 (P = 7 × 10−24), CYP1A2 (P = 1 × 10−23), FGF5 (P = 1 × 10−21), SH2B3 (P = 3 × 10−18), MTHFR (P = 2 × 10−13), c10orf107 (P = 1 × 10−9), ZNF652 (P = 5 × 10−9) and PLCD3 (P = 1 × 10−8) genes. All variants associated with continuous blood pressure were associated with dichotomous hypertension. These associations between common variants and blood pressure and hypertension offer mechanistic insights into the regulation of blood pressure and may point to novel targets for interventions to prevent cardiovascular disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Ezzati, M., Lopez, A.D., Rodgers, A., Vander Hoorn, S. & Murray, C.J. Selected major risk factors and global and regional burden of disease. Lancet 360, 1347–1360 (2002).
Lewington, S., Clarke, R., Qizilbash, N., Peto, R. & Collins, R. Age-specific relevance of usual blood pressure to vascular mortality: a meta-analysis of individual data for one million adults in 61 prospective studies. Lancet 360, 1903–1913 (2002).
The World Health Organization. The World Health Report 2002–Reducing Risks, Promoting Healthy Life (World Health Organization, 2002).
Whelton, P.K. et al. Primary prevention of hypertension: clinical and public health advisory from The National High Blood Pressure Education Program. J. Am. Med. Assoc. 288, 1882–1888 (2002).
Havlik, R.J. et al. Blood pressure aggregation in families. Am. J. Epidemiol. 110, 304–312 (1979).
Lifton, R.P., Gharavi, A.G. & Geller, D.S. Molecular mechanisms of human hypertension. Cell 104, 545–556 (2001).
Ji, W. et al. Rare independent mutations in renal salt handling genes contribute to blood pressure variation. Nat. Genet. 40, 592–599 (2008).
Newhouse, S.J. et al. Haplotypes of the WNK1 gene associate with blood pressure variation in a severely hypertensive population from the British Genetics of Hypertension study. Hum. Mol. Genet. 14, 1805–1814 (2005).
Tobin, M.D. et al. Association of WNK1 gene polymorphisms and haplotypes with ambulatory blood pressure in the general population. Circulation 112, 3423–3429 (2005).
Tobin, M.D. et al. Common variants in genes underlying monogenic hypertension and hypotension and blood pressure in the general population. Hypertension 51, 1658–1664 (2008).
The Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447, 661–678 (2007).
Levy, D. et al. Framingham Heart Study 100K Project: genome-wide associations for blood pressure and arterial stiffness. BMC Med.Genet 8 (Suppl. 1), S3 (2007).
Tobin, M.D., Sheehan, N.A., Scurrah, K.J. & Burton, P.R. Adjusting for treatment effects in studies of quantitative traits: antihypertensive therapy and systolic blood pressure. Stat. Med. 24, 2911–2935 (2005).
Devlin, B. & Roeder, K. Genomic control for association studies. Biometrics 55, 997–1004 (1999).
Pe'er, I., Yelensky, R., Altshuler, D. & Daly, M.J. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet. Epidemiol. 32, 381–385 (2008).
Levy, D. et al. Genome-wide association study of blood pressure and hypertension. Nat. Genet. advance online publication, doi:10.1038/ng.384 (10 May 2009).
Martin, R.M. et al. P450c17 deficiency in Brazilian patients: biochemical diagnosis through progesterone levels confirmed by CYP17 genotyping. J. Clin. Endocrinol. Metab. 88, 5739–5746 (2003).
Geller, D.H., Auchus, R.J., Mendonca, B.B. & Miller, W.L. The genetic and functional basis of isolated 17,20-lyase deficiency. Nat. Genet. 17, 201–205 (1997).
Kluijtmans, L.A. et al. Molecular genetic analysis in mild hyperhomocysteinemia: a common mutation in the methylenetetrahydrofolate reductase gene is a genetic risk factor for cardiovascular disease. Am. J. Hum. Genet. 58, 35–41 (1996).
Sohda, S. et al. Methylenetetrahydrofolate reductase polymorphism and pre-eclampsia. J. Med. Genet. 34, 525–526 (1997).
Qian, X., Lu, Z., Tan, M., Liu, H. & Lu, D. A meta-analysis of association between C677T polymorphism in the methylenetetrahydrofolate reductase gene and hypertension. Eur. J. Hum. Genet. 15, 1239–1245 (2007).
John, S.W. et al. Genetic decreases in atrial natriuretic peptide and salt-sensitive hypertension. Science 267, 679–681 (1995).
Newton-Cheh, C. et al. Association of common variants in NPPA and NPPB with circulating natriuretic peptides and blood pressure. Nat. Genet. 41, 348–353 (2009).
Simon, D.B. et al. Genetic heterogeneity of Bartter's syndrome revealed by mutations in the K+ channel, ROMK. Nat. Genet. 14, 152–156 (1996).
Simon, D.B. et al. Gitelman's variant of Bartter's syndrome, inherited hypokalaemic alkalosis, is caused by mutations in the thiazide-sensitive Na-Cl cotransporter. Nat. Genet. 12, 24–30 (1996).
Daviet, L. et al. Cloning and characterization of ATRAP, a novel protein that interacts with the angiotensin II type 1 receptor. J. Biol. Chem. 274, 17058–17062 (1999).
Suh, P.G. et al. Multiple roles of phosphoinositide-specific phospholipase C isozymes. BMB Rep. 41, 415–434 (2008).
Nebert, D.W. & Dalton, T.P. The role of cytochrome P450 enzymes in endogenous signalling pathways and environmental carcinogenesis. Nat. Rev. Cancer 6, 947–960 (2006).
Sachse, C., Brockmoller, J., Bauer, S. & Roots, I. Functional significance of a C → A polymorphism in intron 1 of the cytochrome P450 CYP1A2 gene tested with caffeine. Br. J. Clin. Pharmacol. 27, 445–449 (1999).
Takebe, A. et al. Microarray analysis of PDGFR alpha+ populations in ES cell differentiation culture identifies genes involved in differentiation of mesoderm and mesenchyme including ARID3b that is essential for development of embryonic mesenchymal cells. Dev. Biol. 293, 25–37 (2006).
Vatner, S.F. FGF induces hypertrophy and angiogenesis in hibernating myocardium. Circ. Res. 96, 705–707 (2005).
Todd, J.A. et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat. Genet. 39, 857–864 (2007).
Hunt, K.A. et al. Newly identified genetic risk variants for celiac disease related to the immune response. Nat. Genet. 40, 395–402 (2008).
Gudbjartsson, D.F. et al. Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat. Genet. 41, 342–347 (2009).
Fitau, J., Boulday, G., Coulon, F., Quillard, T. & Charreau, B. The adaptor molecule Lnk negatively regulates tumor necrosis factor-alpha-dependent VCAM-1 expression in endothelial cells through inhibition of the ERK1 and -2 pathways. J. Biol. Chem. 281, 20148–20159 (2006).
Velazquez, L. et al. Cytokine signaling and hematopoietic homeostasis are disrupted in Lnk-deficient mice. J. Exp. Med. 195, 1599–1611 (2002).
Voight, B.F., Kudaravalli, S., Wen, X. & Pritchard, J.K. A map of recent positive selection in the human genome. PLoS Biol. 4, e72 (2006).
Du, Y.H., Guan, Y.Y., Alp, N.J., Channon, K.M. & Chen, A.F. Endothelium-specific GTP cyclohydrolase I overexpression attenuates blood pressure progression in salt-sensitive low-renin hypertension. Circulation 117, 1045–1054 (2008).
Zheng, J.S. et al. Gene transfer of human guanosine 5′-triphosphate cyclohydrolase I restores vascular tetrahydrobiopterin level and endothelial function in low renin hypertension. Circulation 108, 1238–1245 (2003).
Watanabe, M. et al. Regulation of smooth muscle cell differentiation by AT-rich interaction domain transcription factors Mrf2alpha and Mrf2beta. Circ. Res. 91, 382–389 (2002).
Stamler, J. et al. INTERSALT study findings. Public health and medical care implications. Hypertension 14, 570–577 (1989).
Dyer, A.R., Shipley, M. & Elliott, P. Urinary electrolyte excretion in 24 hours and blood pressure in the INTERSALT Study. I. Estimates of reliability. The INTERSALT Cooperative Research Group. Am. J. Epidemiol. 139, 927–939 (1994).
Hypertension Detection and Follow-up Program Cooperative Group. Variability of blood pressure and the results of screening in the hypertension detection and follow-up program. J. Chronic Dis. 31, 651–667 (1978).
Loos, R.J. et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat. Genet. 40, 768–775 (2008).
Willer, C.J. et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat. Genet. 41, 25–34 (2009).
Kathiresan, S. et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat. Genet. 40, 189–197 (2008).
Li, Y. & Abecasis, G.R. Mach 1.0: rapid haplotype reconstruction and missing genotype inference. Am. J. Hum. Genet. S79, 2290 (2006).
Marchini, J., Howie, B., Myers, S., McVean, G. & Donnelly, P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 39, 906–913 (2007).
Myers, S., Bottolo, L., Freeman, C., McVean, G. & Donnelly, P. A fine-scale map of recombination rates and hotspots across the human genome. Science 310, 321–324 (2005).
Acknowledgements
The authors would like to thank the many colleagues who contributed to collection and phenotypic characterization of the clinical samples, as well as genotyping and analysis of the GWA data. They would also especially like to thank those who agreed to participate in the studies. Major funding for the work described in the paper comes from the following (alphabetically): Academy of Finland (124243, 129322, 129494, 118065), AGAUR (SGR 2005/00577), Albert Påhlsson Research Foundation, Alexander-von-Humboldt Foundation (V-Fokoop-1113183), American Diabetes Association, AstraZeneca AB, AVIS Torino blood donor organization, Barts and The London Charity, Biocenter of University of Oulu, Board of the UMC Utrecht, British Heart Foundation (PG02/128, FS/05/061/19501, SP/04/002), Burroughs Wellcome Fund, CamStrad, Cancer Research United Kingdom, CIBER Epidemiología y Salud Pública, Commissariat à l'Energie Atomique, Compagnia di San Paolo to the ISI Foundation (Torino, Italy), Conservatoire National des Arts et Métiers, Crafoord Foundation, Donovan Family Foundation, Doris Duke Charitable Foundation, Dutch Kidney Foundation (E033), Dutch College of Healthcare Insurance Companies, Dutch Ministry of Health, Dutch Organisation of Health Care Research, ENGAGE (HEALTH-F4-2007-201413), Ernhold Lundströms Research Foundation, Estonian Ministry of Education and Science (0182721s06), EURO-BLCS, European Commission (QLG1-CT-2000-01643, LSHM-CT-2007-037273), European Commission-Europe Against Cancer (AEP/90/05), European Union (FP-6 LSHM-CT-2003-503041, FP-6 LSHM CT 2006 037697), European Society for the Study of Diabetes, Faculty of Biology and Medicine of Lausanne, Switzerland, Fannie E. Rippel Foundation, Finnish Foundation for Cardiovascular Research, FIS (CP05/00290), Fundació Marató Tv3, German Federal Ministry of Education and Research (01ZZ9603, 01ZZ0103, 01ZZ0403, 03ZIK012, 01EZ0874), German National Genome Research Network, German Research Center for Environmental Health, (Neuherberg, Germany), Giorgi-Cavaglieri Foundation, GlaxoSmithKline, Guy's & St. Thomas' NHS Foundation Trust, Health Research and Development Council of the Netherlands (2100.0008, 2100.0042), Helmholtz Zentrum München, Hulda and Conrad Mossfelt Foundation, Institut National de la Recherche Agronomique, Institut National de la Santé et de la Recherche Médicale, Italian Association for Research on Cancer, Italian Ministry of Health (110.1RS97.71), Italian National Research Council, Juvenile Diabetes Research Fund, King Gustaf V and Queen Victoria Foundation, King's College London and King's College Hospital NHS Foundation Trust, Knut and Alice Wallenberg Foundation, Lennart Hanssons Memorial Fund, LK Research Funds, Massachusetts General Hospital Cardiovascular Research Center and Department of Medicine, Medical Faculty of Lund University and Malmö University Hospital, Medical Research Council of the UK (G0000934, G0501942, G9521010D), Medical Research Council-GlaxoSmithKline (85374), MedStar Research Institute, Ministerio de Sanidad y Consumo, Instituto de Salud Carlos III (RD06/0009, CP05/290, PI061254, CIBERESP), Ministry of Cultural Affairs and Social Ministry (Federal State of Mecklenburg-West Pomerania), National Institute for Health Research (NIHR), National Institute for Health Research Cambridge Biomedical Research Centre, Novartis Institute for Biomedical Research, NWO VENI (916.76.170), Province of Utrecht, Region Skane, Siemens Healthcare (Erlangen, Germany), Sigrid Juselius Foundation, Stockholm County Council (562183), Support for Science Funding programme, Swedish Heart and Lung Foundation, Swedish Medical Research Council, Swedish National Research Council, Swedish Research Council (8691), Swiss National Science Foundation (33CSO-122661, 310030-112552, 3100AO-116323/1, PROSPER 3200BO-111362/1, 3233BO-111361/1), UNIL, University of Utrecht, US National Institutes of Health (U01DK062418, K23HL80025, DK062370, DK072193, U54DA021519, 1Z01HG000024, N01AG-916413, N01AG-821336, 263MD916413, 263MD821336, Intramural NIA, R01HL087676, K23HL083102, U54RR020278, R01HL056931, P30ES007033, R01HL087679, RL1MH083268, 263-MA-410953, NO1-AG-1-2109, N01-HD-1-3107), WCRF (98A04, 2000/30), Wellcome Trust (068545/Z/02, 076113/B/04/Z, 079895, 070191/Z/03/Z, 077016/Z/05/Z, WT088885/Z/09/Z).
Author information
Authors and Affiliations
Consortia
Contributions
Author contributions are listed in alphabetical order.
Project conception, design, management: ARYA: M.L.B., C.S.U.; BLSA: L.F.; B58C-T1DGC: D.P.S.; B58C-WTCCC: D.P.S.; BRIGHT: M. Brown, M.C., J.M.C., A. Dominiczak, M.F., P.B.M., N.J.S., J.W.; CoLaus: J.S.B., S. Bergmann, M. Bochud, V.M. (PI), P. Vollenweider (PI), G.W., D.M.W.; DGI: D.A., C.N.-C., L.G.; EPIC-Norfolk-GWAS: I.B., P.D., R.J.F.L., M.S.S., N.J.W., J.H.Z. EPIC-Italy: S. Polidoro, P. Vineis. Fenland Study: R.J.F.L., N.G.F., N.J.W.; Finrisk97: L.P., V.S.; FUSION: R.N.B., M. Boehnke, F.S.C., K.L.M., L.J.S., T.T.V., J.T.; InCHIANTI: S. Bandinelli., L.F.; KORA: A. Döring, C.G., T.I., M. Laan, T.M., E.O., A. Pfeufer, H.E.W. (PI); LOLIPOP: J.C.C., P.E., J.S.K. (PI); MDC-CC: G.B., O.M.; MPP: G.B., O.M.; MIGen: D.A., R.E., S.K., J.M., O.M., C.J.O., V.S., S.M.S., D.S.S.; METSIM: J.K., M. Laakso; NFBC1966: P.E., M.-R.J.; PREVEND: P.E.d.J. (PI), G.N., W.H.v.G.; PROCARDIS: R.C., M.F., A.H., J.F.P., U.S., G.T., H.W. (PI); PROSPECT-EPIC. N.C.O.-M., Y.T.v.d.S.; SardiNIA: E.G.L., D.S.; SHIP: M.D., S.B.F., G.H., R.L., T.R., R.R., U.V., H.V.; SUVIMAX: P.M.; TwinsUK: P.D., T.D.S. (PI); UHP: D.E.G., M.E.N.
Phenotype collection, data management: ARYA: M.L.B., C.S.U.; B58C-T1DGC: D.H., W.L.M., D.P.S.; B58C-WTCCC: D.H., K.P., D.P.S.; BRIGHT: M. Brown, M.C., J.M.C., A. Dominiczak, M.F., P.B.M., N.J.S., J.W.; CoLaus: G.W.; DGI: L.G., O.M.; EPIC-Italy: P. Vineis (PI); Finrisk97: P.J., M.P., V.S.; FUSION: J.T., T.T.V.; KORA: A. Döring, C.G., T.I.; MDC-CC: O.M.; MPP: O.M., P.N.; MIGen: D.A., R.E., S.K., J.M., O.M., C.J.O., S.M.S., D.S.S., V.S.; NFBC1966: A.-L.H., M.-R.J., A. Pouta; PREVEND: P.E.d.J., G.N., P.v.d.H., W.H.v.G.; PROCARDIS: R.C., A.H., U.S., G.T.; PROSPECT-EPIC: N.C.O.-M., Y.T.v.d.S.; SardiNIA: S.S.N., A.S.; SHIP: M.D., R.L., R.R., H.V.; SUVIMAX: P.G., S.H.; TwinsUK: F.M.W.; UHP: D.E.G., M.E.N.
Genome-wide, validation genotyping: B58C-T1DGC: W.L.M.; B58C-WTCCC: W.L.M.; DGI: D.A., O.M., M.O.-M.; EPIC-Norfolk-GWAS: I.B., P.D., N.J.W., J.H.Z.; EPIC-Norfolk-replication: S.A.B., K.-T.K., R.J.F.L., R.N.L., N.J.W.; EPIC-Italy: A.A., A.D.G., S.G., G.M., V.R.; Finrisk97: G.J.C., C.N.-C.; FUSION: L.L.B., M.A.M.; KORA: T.I., T.M., E.O., A. Pfeufer; MDC-CC: O.M., M.O.-M.; MPP: O.M., M.O.-M.; NFBC1966: P.E., N.B.F., M.-R.J., M.I.M., L.P. ; PREVEND: G.N., P.v.d.H.; W.H.v.G.; PROCARDIS: S.C.H., G.M.L., A.-C.S.; SardiNIA: M.U.; SHIP: F.E., G.H., A.T., U.V.; SUVIMAX: I.G.G., S.C.H., G.M.L., D.Z.; TwinsUK: P.D., N.S.
Data analysis: BLSA: T.T.; B58C-T1DGC: D.H., S.H., D.P.S.; B58C-WTCCC: P.R.B., D.H., K.P., D.P.S, M.D.T.; BRIGHT: S.J.N., C.W., E.Z.; CoLaus: S. Bergmann, M. Bochud, T.J., N.L., K.S., X.Y., DGI: O.M., C.N.-C., M.O.-M., B.F.V.; EPIC-Norfolk-GWAS: R.J.F.L., J.H.Z.; EPIC-Norfolk-replication: S.A.B., K.-T.K., R.J.F.L., R.N.L., N.J.W.; EPIC-Italy: S.G., G.M., S. Panico, S. Polidoro, F.R., C.S., P. Vineis; Fenland Study: J.L.; Finrisk97: C.N.-C.; FUSION: A.U.J., L.J.S., H.M.S., C.J.W.; InCHIANTI: T.T.; KORA: S.E., C.G., M. Laan, E.O.; LOLIPOP: J.C.C.; MDC-CC: O.M., M.O.-M.; MPP: O.M., M.O.-M.; MIGen: R.E., G.L., I.S., B.F.V.; NFBC1966: L.C., P.F.O.; PREVEND: H.S., P.v.d.H.; PROCARDIS: M.F., A.G., J.F.P.; SardiNIA: V.G., S.S., P.S.; SHIP: F.E., G.H., A.T., U.V.; SUVIMAX: S.C.H., T.J., P.M.; TwinsUK: N.S., F.Z., G.Z.
Analysis group: G.R.A., M.C., V.G., T.J., P.B.M., C.N.-C., M.D.T., L.V.W.
Writing group: G.R.A., M.C., P.E., V.G., T.J., P.B.M., C.N.-C., M.D.T.
Corresponding authors
Ethics declarations
Competing interests
The following authors declare the following potential conflicts of interest: N.L., V.M., K.S., D.M.W. and X.Y. are all full-time employees at GlaxoSmithKline. P.Vollenweider and G.W received financial support from GlaxoSmithKline to assemble the CoLaus study. No other authors reported conflicts of interest.
Additional information
A full list of authors is provided in the Supplementary Note.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1 and 2, Supplementary Tables 1–4, Supplementary Methods and Supplementary Note (PDF 2563 kb)
Rights and permissions
About this article
Cite this article
Newton-Cheh, C., Johnson, T., Gateva, V. et al. Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet 41, 666–676 (2009). https://doi.org/10.1038/ng.361
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.361
This article is cited by
-
New insights into the nutritional genomics of adult-onset riboflavin-responsive diseases
Nutrition & Metabolism (2023)
-
Mutated CYP17A1 promotes atherosclerosis and early-onset coronary artery disease
Cell Communication and Signaling (2023)
-
Leveraging molecular quantitative trait loci to comprehend complex diseases/traits from the omics perspective
Human Genetics (2023)
-
Severe Pulmonary Arterial Hypertension in Healthy Young Infants: Single Center Experience
Indian Pediatrics (2023)
-
A deep learning model for early risk prediction of heart failure with preserved ejection fraction by DNA methylation profiles combined with clinical features
Clinical Epigenetics (2022)