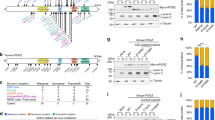
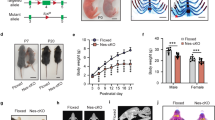
Abstract
Numerous genes are associated with neurodevelopmental disorders such as intellectual disability and autism spectrum disorder (ASD), but their dysfunction is often poorly characterized. Here we identified dominant mutations in the gene encoding the transcriptional repressor and MeCP2 interactor switch-insensitive 3 family member A (SIN3A; chromosome 15q24.2) in individuals who, in addition to mild intellectual disability and ASD, share striking features, including facial dysmorphisms, microcephaly and short stature. This phenotype is highly related to that of individuals with atypical 15q24 microdeletions, linking SIN3A to this microdeletion syndrome. Brain magnetic resonance imaging showed subtle abnormalities, including corpus callosum hypoplasia and ventriculomegaly. Intriguingly, in vivo functional knockdown of Sin3a led to reduced cortical neurogenesis, altered neuronal identity and aberrant corticocortical projections in the developing mouse brain. Together, our data establish that haploinsufficiency of SIN3A is associated with mild syndromic intellectual disability and that SIN3A can be considered to be a key transcriptional regulator of cortical brain development.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Accession codes
References
Lugtenberg, D. et al. De novo loss-of-function mutations in WAC cause a recognizable intellectual disability syndrome and learning deficits in Drosophila. Eur. J. Hum. Genet. http://dx.doi.org/10.1038/ejhg.2015.282 (2016).
Willemsen, M.H. & Kleefstra, T. Making headway with genetic diagnostics of intellectual disabilities. Clin. Genet. 85, 101–110 (2014).
Willemsen, M.H. et al. Chromosome 1p21.3 microdeletions comprising DPYD and MIR137 are associated with intellectual disability. J. Med. Genet. 48, 810–818 (2011).
Samocha, K.E. et al. A framework for the interpretation of de novo mutation in human disease. Nat. Genet. 46, 944–950 (2014).
Coe, B.P. et al. Refining analyses of copy number variation identifies specific genes associated with developmental delay. Nat. Genet. 46, 1063–1071 (2014).
Gilissen, C. et al. Genome sequencing identifies major causes of severe intellectual disability. Nature 511, 344–347 (2014).
Jansen, S. et al. De novo loss-of-function mutations in X-linked SMC1A cause severe ID and therapy resistant epilepsy in females: expanding the phenotypic spectrum. Clin. Genet. http://dx.doi.org/10.1111/cge.12729 (2016).
Magoulas, P.L. & El-Hattab, A.W. Chromosome 15q24 microdeletion syndrome. Orphanet J. Rare Dis. 7, 2 (2012).
Mefford, H.C. et al. Further clinical and molecular delineation of the 15q24 microdeletion syndrome. J. Med. Genet. 49, 110–118 (2012).
Andrieux, J. et al. Genotype–phenotype correlation in four 15q24 deleted patients identified by array-CGH. Am. J. Med. Genet. A. 149A, 2813–2819 (2009).
Borrell, V. & Reillo, I. Emerging roles of neural stem cells in cerebral cortex development and evolution. Dev. Neurobiol. 72, 955–971 (2012).
Fish, J.L., Dehay, C., Kennedy, H. & Huttner, W.B. Making bigger brains—the evolution of neural-progenitor-cell division. J. Cell Sci. 121, 2783–2793 (2008).
Laguesse, S., Peyre, E. & Nguyen, L. Progenitor genealogy in the developing cerebral cortex. Cell Tissue Res. 359, 17–32 (2015).
Schubert, D., Martens, G.J. & Kolk, S.M. Molecular underpinnings of prefrontal cortex development in rodents provide insights into the etiology of neurodevelopmental disorders. Mol. Psychiatry 20, 795–809 (2015).
Bielen, H. & Houart, C. The Wnt cries many: Wnt regulation of neurogenesis through tissue patterning, proliferation, and asymmetric cell division. Dev. Neurobiol. 74, 772–780 (2014).
LaMonica, B.E., Lui, J.H., Wang, X. & Kriegstein, A.R. OSVZ progenitors in the human cortex: an updated perspective on neurodevelopmental disease. Curr. Opin. Neurobiol. 22, 747–753 (2012).
Dannenberg, J.H. et al. mSin3A corepressor regulates diverse transcriptional networks governing normal and neoplastic growth and survival. Genes Dev. 19, 1581–1595 (2005).
Gallagher, S.J. et al. Distinct requirements for Sin3a in perinatal male gonocytes and differentiating spermatogonia. Dev. Biol. 373, 83–94 (2013).
McDonel, P., Demmers, J., Tan, D.W., Watt, F. & Hendrich, B.D. Sin3a is essential for the genome integrity and viability of pluripotent cells. Dev. Biol. 363, 62–73 (2012).
Gajan, A., Barnes, V.L., Liu, M., Saha, N. & Pile, L.A. The histone demethylase dKDM5/LID interacts with the SIN3 histone deacetylase complex and shares functional similarities with SIN3. Epigenetics Chromatin 9, 4 (2016).
Saha, N., Liu, M., Gajan, A. & Pile, L.A. Genome-wide studies reveal novel and distinct biological pathways regulated by SIN3 isoforms. BMC Genomics 17, 111 (2016).
Cowley, S.M. et al. The mSin3A chromatin-modifying complex is essential for embryogenesis and T-cell development. Mol. Cell. Biol. 25, 6990–7004 (2005).
Nascimento, E.M. et al. The opposing transcriptional functions of Sin3a and c-Myc are required to maintain tissue homeostasis. Nat. Cell Biol. 13, 1395–1405 (2011).
Bansal, N., David, G., Farias, E. & Waxman, S. Emerging roles of epigenetic regulator Sin3 in cancer. Adv. Cancer Res. 130, 113–135 (2016).
Schoch, H. & Abel, T. Transcriptional co-repressors and memory storage. Neuropharmacology 80, 53–60 (2014).
Kadamb, R., Mittal, S., Bansal, N., Batra, H. & Saluja, D. Sin3: insight into its transcription regulatory functions. Eur. J. Cell Biol. 92, 237–246 (2013).
Huang, Y., Myers, S.J. & Dingledine, R. Transcriptional repression by REST: recruitment of Sin3A and histone deacetylase to neuronal genes. Nat. Neurosci. 2, 867–872 (1999).
Kyrylenko, S., Korhonen, P., Kyrylenko, O., Roschier, M. & Salminen, A. Expression of transcriptional repressor proteins mSin3A and 3B during aging and replicative senescence. Biochem. Biophys. Res. Commun. 275, 455–459 (2000).
Kolk, S.M., de Mooij-Malsen, A.J. & Martens, G.J. Spatiotemporal molecular approach of in utero electroporation to functionally decipher endophenotypes in neurodevelopmental disorders. Front. Mol. Neurosci. 4, 37 (2011).
Swaminathan, A. & Pile, L.A. Regulation of cell proliferation and wing development by Drosophila SIN3 and String. Mech. Dev. 127, 96–106 (2010).
Miller, J.A. et al. Transcriptional landscape of the prenatal human brain. Nature 508, 199–206 (2014).
Lein, E.S. et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176 (2007).
Laird, A.R., Lancaster, J.L. & Fox, P.T. BrainMap: the social evolution of a human brain mapping database. Neuroinformatics 3, 65–78 (2005).
Fox, P.T. et al. BrainMap taxonomy of experimental design: description and evaluation. Hum. Brain Mapp. 25, 185–198 (2005).
Fox, P.T. & Lancaster, J.L. Mapping context and content: the BrainMap model. Nat. Rev. Neurosci. 3, 319–321 (2002).
Götz, M. & Huttner, W.B. The cell biology of neurogenesis. Nat. Rev. Mol. Cell Biol. 6, 777–788 (2005).
Noctor, S.C., Martínez-Cerdeño, V., Ivic, L. & Kriegstein, A.R. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat. Neurosci. 7, 136–144 (2004).
Noctor, S.C., Martínez-Cerdeño, V. & Kriegstein, A.R. Distinct behaviors of neural stem and progenitor cells underlie cortical neurogenesis. J. Comp. Neurol. 508, 28–44 (2008).
Evsyukova, I., Plestant, C. & Anton, E.S. Integrative mechanisms of oriented neuronal migration in the developing brain. Annu. Rev. Cell Dev. Biol. 29, 299–353 (2013).
Molyneaux, B.J., Arlotta, P., Menezes, J.R. & Macklis, J.D. Neuronal subtype specification in the cerebral cortex. Nat. Rev. Neurosci. 8, 427–437 (2007).
Leone, D.P., Srinivasan, K., Chen, B., Alcamo, E. & McConnell, S.K. The determination of projection neuron identity in the developing cerebral cortex. Curr. Opin. Neurobiol. 18, 28–35 (2008).
Shoemaker, L.D. & Arlotta, P. Untangling the cortex: advances in understanding specification and differentiation of corticospinal motor neurons. BioEssays 32, 197–206 (2010).
Namba, T. et al. Pioneering axons regulate neuronal polarization in the developing cerebral cortex. Neuron 81, 814–829 (2014).
Zolessi, F.R., Poggi, L., Wilkinson, C.J., Chien, C.B. & Harris, W.A. Polarization and orientation of retinal ganglion cells in vivo. Neural Dev. 1, 2 (2006).
Hatanaka, Y. et al. Distinct roles of neuropilin 1 signaling for radial and tangential extension of callosal axons. J. Comp. Neurol. 514, 215–225 (2009).
Nan, X. et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393, 386–389 (1998).
Boeke, J., Ammerpohl, O., Kegel, S., Moehren, U. & Renkawitz, R. The minimal repression domain of MBD2b overlaps with the methyl-CpG-binding domain and binds directly to Sin3A. J. Biol. Chem. 275, 34963–34967 (2000).
Baltus, G.A., Kowalski, M.P., Tutter, A.V. & Kadam, S. A positive regulatory role for the mSin3A–HDAC complex in pluripotency through Nanog and Sox2. J. Biol. Chem. 284, 6998–7006 (2009).
Liang, J. et al. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat. Cell Biol. 10, 731–739 (2008).
Rampalli, S., Pavithra, L., Bhatt, A., Kundu, T.K. & Chattopadhyay, S. Tumor suppressor SMAR1 mediates cyclin D1 repression by recruitment of the SIN3/histone deacetylase 1 complex. Mol. Cell. Biol. 25, 8415–8429 (2005).
Ji, Q. et al. CRL4B interacts with and coordinates the SIN3A–HDAC complex to repress CDKN1A and drive cell cycle progression. J. Cell Sci. 127, 4679–4691 (2014).
Pollock, A., Bian, S., Zhang, C., Chen, Z. & Sun, T. Growth of the developing cerebral cortex is controlled by microRNA-7 through the p53 pathway. Cell Reports 7, 1184–1196 (2014).
van Oevelen, C. et al. A role for mammalian Sin3 in permanent gene silencing. Mol. Cell 32, 359–370 (2008).
Suzuki, D.E., Ariza, C.B., Porcionatto, M.A. & Okamoto, O.K. Upregulation of E2F1 in cerebellar neuroprogenitor cells and cell cycle arrest during postnatal brain development. In Vitro Cell. Dev. Biol. Anim. 47, 492–499 (2011).
Kim, Y. et al. Activation of Cdk2–pRB–E2F1 cell cycle pathway by repeated electroconvulsive shock in the rat frontal cortex. Biol. Psychiatry 57, 107–109 (2005).
Kleefstra, T., Schenck, A., Kramer, J.M. & van Bokhoven, H. The genetics of cognitive epigenetics. Neuropharmacology 80, 83–94 (2014).
Chahrour, M. & Zoghbi, H.Y. The story of Rett syndrome: from clinic to neurobiology. Neuron 56, 422–437 (2007).
Damen, D. & Heumann, R. MeCP2 phosphorylation in the brain: from transcription to behavior. Biol. Chem. 394, 1595–1605 (2013).
Krishnan, K. et al. MeCP2 regulates the timing of critical period plasticity that shapes functional connectivity in primary visual cortex. Proc. Natl. Acad. Sci. USA 112, E4782–E4791 (2015).
Kishi, N. & Macklis, J.D. MECP2 is progressively expressed in post-migratory neurons and is involved in neuronal maturation rather than cell fate decisions. Mol. Cell. Neurosci. 27, 306–321 (2004).
Zhang, W., Peterson, M., Beyer, B., Frankel, W.N. & Zhang, Z.W. Loss of MeCP2 from forebrain excitatory neurons leads to cortical hyperexcitation and seizures. J. Neurosci. 34, 2754–2763 (2014).
Chao, H.T., Zoghbi, H.Y. & Rosenmund, C. MeCP2 controls excitatory synaptic strength by regulating glutamatergic synapse number. Neuron 56, 58–65 (2007).
Dani, V.S. et al. Reduced cortical activity due to a shift in the balance between excitation and inhibition in a mouse model of Rett syndrome. Proc. Natl. Acad. Sci. USA 102, 12560–12565 (2005).
Vignoli, A. et al. Correlations between neurophysiological, behavioral, and cognitive function in Rett syndrome. Epilepsy Behav. 17, 489–496 (2010).
Samuelsson, L., Zagoras, T. & Hafström, M. Inherited 15q24 microdeletion syndrome in twins and their father with phenotypic variability. Eur. J. Med. Genet. 58, 111–115 (2015).
Silverstein, R.A. & Ekwall, K. Sin3: a flexible regulator of global gene expression and genome stability. Curr. Genet. 47, 1–17 (2005).
Paul, L.K. Developmental malformation of the corpus callosum: a review of typical callosal development and examples of developmental disorders with callosal involvement. J. Neurodev. Disord. 3, 3–27 (2011).
Guerrini, R. & Dobyns, W.B. Malformations of cortical development: clinical features and genetic causes. Lancet Neurol. 13, 710–726 (2014).
Yu, T.W. et al. Mutations in WDR62, encoding a centrosome-associated protein, cause microcephaly with simplified gyri and abnormal cortical architecture. Nat. Genet. 42, 1015–1020 (2010).
Bilgüvar, K. et al. Whole-exome sequencing identifies recessive WDR62 mutations in severe brain malformations. Nature 467, 207–210 (2010).
Chen, J.F. et al. Microcephaly disease gene Wdr62 regulates mitotic progression of embryonic neural stem cells and brain size. Nat. Commun. 5, 3885 (2014).
Xu, D., Zhang, F., Wang, Y., Sun, Y. & Xu, Z. Microcephaly-associated protein WDR62 regulates neurogenesis through JNK1 in the developing neocortex. Cell Reports 6, 104–116 (2014).
Nicholas, A.K. et al. WDR62 is associated with the spindle pole and is mutated in human microcephaly. Nat. Genet. 42, 1010–1014 (2010).
Huang, T.N. et al. Tbr1 haploinsufficiency impairs amygdalar axonal projections and results in cognitive abnormality. Nat. Neurosci. 17, 240–247 (2014).
Kolk, S.M., Whitman, M.C., Yun, M.E., Shete, P. & Donoghue, M.J. A unique subpopulation of Tbr1-expressing deep layer neurons in the developing cerebral cortex. Mol. Cell. Neurosci. 32, 200–214 (2006).
Bedogni, F. et al. Tbr1 regulates regional and laminar identity of postmitotic neurons in developing neocortex. Proc. Natl. Acad. Sci. USA 107, 13129–13134 (2010).
Palumbo, O. et al. TBR1 is the candidate gene for intellectual disability in patients with a 2q24.2 interstitial deletion. Am. J. Med. Genet. A. 164A, 828–833 (2014).
Deriziotis, P. et al. De novo TBR1 mutations in sporadic autism disrupt protein functions. Nat. Commun. 5, 4954 (2014).
O'Roak, B.J. et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science 338, 1619–1622 (2012).
Kolk, S.M. et al. Semaphorin 3F is a bifunctional guidance cue for dopaminergic axons and controls their fasciculation, channeling, rostral growth, and intracortical targeting. J. Neurosci. 29, 12542–12557 (2009).
McDonel, P., Costello, I. & Hendrich, B. Keeping things quiet: roles of NuRD and Sin3 co-repressor complexes during mammalian development. Int. J. Biochem. Cell Biol. 41, 108–116 (2009).
Lin, T. et al. p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat. Cell Biol. 7, 165–171 (2005).
Yuan, B., Latek, R., Hossbach, M., Tuschl, T. & Lewitter, F. siRNA Selection Server: an automated siRNA oligonucleotide prediction server. Nucleic Acids Res. 32, W130–W134 (2004).
North, H.A. et al. Promotion of proliferation in the developing cerebral cortex by EphA4 forward signaling. Development 136, 2467–2476 (2009).
Acknowledgements
We thank laboratory members and colleagues for critically reading this manuscript and members of the various laboratories for helpful editing and discussions. We express thanks to W. Hendriks (Radboud University) for sharing plasmids and N. Nadif Kasri (Radboud University Medical Center) for kindly providing antibody to mouse Ki-67. We are grateful for the mouse Sin3a cDNA clone from R. Floyd and B.D. Hendrich (Wellcome Trust Centre for Stem Cell Research and MRC Centre for Stem Cell Biology and Regenerative Medicine). We thank the Radboud Institute for Molecular Life Sciences microscopy platform (see URLs) for excellent support and maintenance of the equipment. We are grateful to the families and subjects participating in this study for their involvement. This work was supported by funding from Science without Borders, CAPES-Brasil (BEX 12044/13-0) to T.C.D.D., complemented by extra support from the Educational Institute of Biosciences at Radboud University, by grants from the Netherlands Organization for Health Research and Development, ZonMw (grant 907-00-365) to T.K., by the Dutch Brain Foundation (HsN F2014(1)-16) to J.E.V. and by the German Ministry of Research and Education (grants 01GS08164, 01GS08167 and 01GS08163 German Mental Retardation Network) to H.E. and T.S., as part of the National Genome Research Network.
Author information
Authors and Affiliations
Contributions
The study was designed and directed by T.K. and S.M.K. Patient ascertainment and recruitment were carried out by T.K., M.H.W., H.E.V.-K., C.M.A.v.R.-A., D.V., J.S.K.W.-R., M.V., A.D., J.S., P.R., N.F., K.C., S.A.d.M., C.L.C. and H.G.B. Microarray analyses, DNA sequencing, validation and genotyping were carried out and interpreted by W.M.N., T.S., A.M.Z., H.E. and J.S.W. C.G. was responsible for the bioinformatics of human genetic data analyses. R.P., T.C.D.D., N.H.M.v.B. and E.J.R.J. performed the in vitro functional assays, cloning and mouse experiments. J.E.V. interpreted the in vitro functional assays, cloning and mouse experiments. J.A.v.H. was invaluable in mouse care. The manuscript was written by J.S.W., M.H.W., T.C.D.D., G.J.M.M., T.K. and S.M.K., with all authors refining and approving the final version of it.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Schematic of the deleted regions on chromosome 15q24 in individuals 1– 4 and two previously reported individuals with a 15q24 microdeletion.
The chromosomal 15q24 region contains several segmental duplication blocks, including breakpoints A, B, C, D and E (breakpoints C and D are indicated), which are thought to predispose to the occurrence of deletions and duplications in this region by non-allelic homologous recombination (NAHR) during meiosis. Different deletions mediated by different combinations of segmental duplication blocks were previously reported. In addition, some individuals have an atypical deletion in which one or both of the breakpoints are not located in a segmental duplication block. Consequently, the clinical features of the 15q24 microdeletion syndrome are heterogeneous. Genotype–phenotype studies of individuals with typical overlapping deletions have suggested the 1.1-Mb region between segmental duplication blocks B and C (72.2–73.3 Mb, hg18; 74.4–75.5 Mb, hg19) as the critical region for the core phenotype. However, Mefford et al. reported two small de novo deletions only involving the region between breakpoints C and D. These two individuals shared only five genes in the deleted region: PTPN9, SIN3A, MAN2C, NEIL1 and COMMD4. The phenotype of these individuals was reported to be milder with less pronounced speech delay than in individuals with the larger deletions between breakpoints B and C.
Supplementary Figure 2 Developmental transcriptome and LMD microarray analysis show high expression levels of SIN3A in proliferative regions of human developing cortex.
(a) Overview of the online database of the Allen Institute for Brain Science showing RNA-seq data across developmental stages (from 4–7 weeks post-conception (wpc) into adulthood) of human brain development showing low to moderate expression of SIN3A across development in various brain structures. (b) Detail of the expression levels of SIN3A in cortical regions 9 wpc, with a higher expression level in temporal neocortex (TGx). (c) Detail of the expression levels of SIN3A in cortical regions 9 wpc, with a higher expression level in the primary motor-sensory cortex (M1C-S1C). (d) Overview of the online database of the Allen Institute for Brain Science showing LMD microarray data across developmental stages (from 4–7 wpc into adulthood) of human brain development showing low to moderate expression of SIN3A 21 wpc in the ventricular zone (VZ) of the posterior frontal cortex (motor cortex). (e) Comparison of the expression levels of SIN3A in deeper cortical regions (e.g., VZ) and more superficial cortical regions (e.g., marginal zone, MZ) 21 wpc. Details on the complete SIN3A transcriptome profile can be found at http://www.brainspan.org/.
Supplementary Figure 3 Protein expression of Sin3a over time and Nissl validation.
(a) Immunostaining for Sin3a (green) at E14.5, E16.5, E18.5, P7, P14 and P21 and counterstaining with fluorescent Nissl (blue). (b) Coronal sections showing the somatosensory cortical area (S1) of E14.5 (left) and E16.5 (right) mouse brains with immunostaining for Sin3a (green) and Ki-67 (red); sections were counterstained with fluorescent Nissl (blue). Arrows and insets show colocalization (yellow). (c) Representative images of Nissl staining of a control (Ctrl)-electroporated cortical area (left) and an shSin3a-electroporated cortical area (right) immunostained for GFP (green) and counterstained with fluorescent Nissl (blue) flanked by a black-and-white image of Nissl staining (asterisks in the cell sparser area).
Supplementary Figure 4 Validation of Sin3a knockdown at the mRNA level.
(a) Relative expression levels (percentage) of Sin3a (tested with two primer pairs (PP1 and PP2; two shades of grey) in comparison to β-actin (black) in mouse brain at P35 and N2a cells. (b) Normalized expression levels of Sin3a mRNA in N2a cells transfected with two siRNAs targeting Sin3a mRNA (siSin3a-ex13 and siSin3a-ex16), two scrambled siRNAs (sc-siSin3a-ex13 and sc-siSin3a-ex16) in comparison to mock (no construct) as a control determined by qPCR using two primer pairs (PP1, black; PP2, grey). (c–g) Schematic of the constructs containing the shRNA for Sin3a exon 13 and exon 16 with accompanying scrambled constructs and the pCAB expression vector with the shRNA-insensitive mSin3a*. (h–j) Normalized mRNA expression levels of Cdkn1a, Mecp2 and Ccnd1 (n = 3 biological replicates) did not change 48 h after knockdown of Sin3a. Data are presented as normalized mean transcript levels ± s.e.m. Student’s t test.
Supplementary Figure 5 Validation of in vivo Sin3a knockdown at the protein level.
(a) Representative images are shown of the electroporated (shSin3a-ex13, green) area double labeled with Sin3a (red) and counterstained with fluorescent Nissl (blue; left). At the site of electroporation with shRNA, Sin3a protein levels are downregulated (white arrows; right). (b) Representative image of an electroporated (shSin3a-ex13, green) area double labeled with cleaved caspase 3 (CC3) showing an apoptotic cell double labeled with GFP (arrowhead) and in the vicinity of GFP-labeled cells (arrow). (c) Positive control showing an area (septal area E17.5) positive for CC3. (d) Quantification of the number of CC3-positive cells in the electroporated area; n = 4 for Ctrl, n = 5 for shSin3a and n = 2 for shSin3a + mSin3a*. Data are presented as the number of cells per mm2 ± s.e.m. One-way ANOVA (α = 0.05). (e) Quantification of the number of GFP+CC3+ cells in the electroporated area as an indication for cell-autonomous effects; n = 4 for Ctrl, n = 5 for shSin3a and n = 2 for shSin3a + mSin3a*. Data are presented as the number of cells per mm2 ± s.e.m. One-way ANOVA (α = 0.05).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5, Supplementary Tables 1 and 2, and Supplementary Note. (PDF 1186 kb)
Rights and permissions
About this article
Cite this article
Witteveen, J., Willemsen, M., Dombroski, T. et al. Haploinsufficiency of MeCP2-interacting transcriptional co-repressor SIN3A causes mild intellectual disability by affecting the development of cortical integrity. Nat Genet 48, 877–887 (2016). https://doi.org/10.1038/ng.3619
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3619
This article is cited by
-
ZSWIM4 regulates embryonic patterning and BMP signaling by promoting nuclear Smad1 degradation
EMBO Reports (2024)
-
Novel phenotype of SIN3A-related disorder diagnosed in adulthood with multi-system involvement
European Journal of Human Genetics (2024)
-
Sin3a drives mesenchymal-to-epithelial transition through cooperating with Tet1 in somatic cell reprogramming
Stem Cell Research & Therapy (2022)
-
Development of prefrontal cortex
Neuropsychopharmacology (2022)
-
Characterization of gene expression profiles in the mouse brain after 35 days of spaceflight mission
npj Microgravity (2022)