Abstract
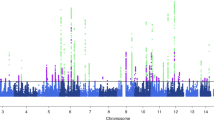
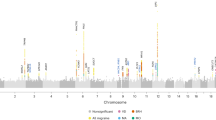
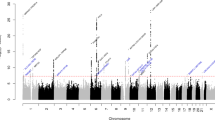
Migraine is a debilitating neurological disorder affecting around one in seven people worldwide, but its molecular mechanisms remain poorly understood. There is some debate about whether migraine is a disease of vascular dysfunction or a result of neuronal dysfunction with secondary vascular changes. Genome-wide association (GWA) studies have thus far identified 13 independent loci associated with migraine. To identify new susceptibility loci, we carried out a genetic study of migraine on 59,674 affected subjects and 316,078 controls from 22 GWA studies. We identified 44 independent single-nucleotide polymorphisms (SNPs) significantly associated with migraine risk (P < 5 × 10−8) that mapped to 38 distinct genomic loci, including 28 loci not previously reported and a locus that to our knowledge is the first to be identified on chromosome X. In subsequent computational analyses, the identified loci showed enrichment for genes expressed in vascular and smooth muscle tissues, consistent with a predominant theory of migraine that highlights vascular etiologies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Change history
18 July 2016
In the version of this article initially published online, the affiliations for Bertram Muller-Myhsok and Patricia Pozo-Rosich were incorrect or incomplete. These errors have been corrected for the print, PDF and HTML versions of this article.
References
Vos, T. et al. Years lived with disability (YLDs) for 1160 sequelae of 289 diseases and injuries 1990–2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 380, 2163–2196 (2012).
Vos, T. et al. Global, regional, and national incidence, prevalence, and years lived with disability for 301 acute and chronic diseases and injuries in 188 countries, 1990–2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet 386, 743–800 (2015).
Gustavsson, A. et al. Cost of disorders of the brain in Europe 2010. Eur. Neuropsychopharmacol. 21, 718–779 (2011).
Pietrobon, D. & Striessnig, J. Neurological diseases: neurobiology of migraine. Nat. Rev. Neurosci. 4, 386–398 (2003).
Tfelt-Hansen, P.C. & Koehler, P.J. One hundred years of migraine research: major clinical and scientific observations from 1910 to 2010. Headache 51, 752–778 (2011).
Headache Classification Committee of the International Headache Society (IHS) The International Classification of Headache Disorders, 3rd edition (beta version). Cephalalgia 33, 629–808 (2013).
Polderman, T.J.C. et al. Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nat. Genet. 47, 702–709 (2015).
Anttila, V. et al. Genome-wide association study of migraine implicates a common susceptibility variant on 8q22.1. Nat. Genet. 42, 869–873 (2010).
Chasman, D.I. et al. Genome-wide association study reveals three susceptibility loci for common migraine in the general population. Nat. Genet. 43, 695–698 (2011).
Freilinger, T. et al. Genome-wide association analysis identifies susceptibility loci for migraine without aura. Nat. Genet. 44, 777–782 (2012).
Anttila, V. et al. Genome-wide meta-analysis identifies new susceptibility loci for migraine. Nat. Genet. 45, 912–917 (2013).
Ophoff, R.A. et al. Familial hemiplegic migraine and episodic ataxia type-2 are caused by mutations in the Ca2+ channel gene CACNL1A4. Cell 87, 543–552 (1996).
De Fusco, M. et al. Haploinsufficiency of ATP1A2 encoding the Na+/K+ pump α2 subunit associated with familial hemiplegic migraine type 2. Nat. Genet. 33, 192–196 (2003).
Dichgans, M. et al. Mutation in the neuronal voltage-gated sodium channel SCN1A in familial hemiplegic migraine. Lancet 366, 371–377 (2005).
Nyholt, D.R. et al. A high-density association screen of 155 ion transport genes for involvement with common migraine. Hum. Mol. Genet. 17, 3318–3331 (2008).
1000 Genomes Project Consortium et al. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 (2012).
Chasman, D.I. et al. Selectivity in genetic association with sub-classified migraine in women. PLoS Genet. 10, e1004366 (2014).
Han, B. & Eskin, E. Random-effects model aimed at discovering associations in meta-analysis of genome-wide association studies. Am. J. Hum. Genet. 88, 586–598 (2011).
Morton, M.J., Abohamed, A., Sivaprasadarao, A. & Hunter, M. pH sensing in the two-pore domain K+ channel, TASK2. Proc. Natl. Acad. Sci. USA 102, 16102–16106 (2005).
Ramachandran, R. et al. TRPM8 activation attenuates inflammatory responses in mouse models of colitis. Proc. Natl. Acad. Sci. USA 110, 7476–7481 (2013).
Hanna, M.G. Genetic neurological channelopathies. Nat. Clin. Pract. Neurol. 2, 252–263 (2006).
Kraev, A. et al. Molecular cloning of a third member of the potassium-dependent sodium-calcium exchanger gene family, NCKX3. J. Biol. Chem. 276, 23161–23172 (2001).
Ismailov, I.I. et al. A biologic function for an 'orphan' messenger: D-myo-inositol 3,4,5,6-tetrakisphosphate selectively blocks epithelial calcium-activated chloride channels. Proc. Natl. Acad. Sci. USA 93, 10505–10509 (1996).
De Bock, M. et al. Connexin channels provide a target to manipulate brain endothelial calcium dynamics and blood-brain barrier permeability. J. Cereb. Blood Flow Metab. 31, 1942–1957 (2011).
Kathiresan, S. et al. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat. Genet. 41, 334–341 (2009).
Debette, S. et al. Common variation in PHACTR1 is associated with susceptibility to cervical artery dissection. Nat. Genet. 47, 78–83 (2015).
Law, C. et al. Clinical features in a family with an R460H mutation in transforming growth factor β receptor 2 gene. J. Med. Genet. 43, 908–916 (2006).
Bown, M.J. et al. Abdominal aortic aneurysm is associated with a variant in low-density lipoprotein receptor-related protein 1. Am. J. Hum. Genet. 89, 619–627 (2011).
Arndt, A.K. et al. Fine mapping of the 1p36 deletion syndrome identifies mutation of PRDM16 as a cause of cardiomyopathy. Am. J. Hum. Genet. 93, 67–77 (2013).
Fujimura, M. et al. Genetics and biomarkers of Moyamoya disease: significance of RNF213 as a susceptibility gene. J. Stroke 16, 65–72 (2014).
McElhinney, D.B. et al. Analysis of cardiovascular phenotype and genotype-phenotype correlation in individuals with a JAG1 mutation and/or Alagille syndrome. Circulation 106, 2567–2574 (2002).
Bezzina, C.R. et al. Common variants at SCN5A–SCN10A and HEY2 are associated with Brugada syndrome, a rare disease with high risk of sudden cardiac death. Nat. Genet. 45, 1044–1049 (2013).
Sinner, M.F. et al. Integrating genetic, transcriptional, and functional analyses to identify five novel genes for atrial fibrillation. Circulation 130, 1225–1235 (2014).
Neale, B.M. et al. Genome-wide association study of advanced age-related macular degeneration identifies a role of the hepatic lipase gene (LIPC). Proc. Natl. Acad. Sci. USA 107, 7395–7400 (2010).
Desch, M. et al. IRAG determines nitric oxide- and atrial natriuretic peptide-mediated smooth muscle relaxation. Cardiovasc. Res. 86, 496–505 (2010).
Lang, N.N., Luksha, L., Newby, D.E. & Kublickiene, K. Connexin 43 mediates endothelium-derived hyperpolarizing factor-induced vasodilatation in subcutaneous resistance arteries from healthy pregnant women. Am. J. Physiol. Heart Circ. Physiol. 292, H1026–H1032 (2007).
Dong, H., Jiang, Y., Triggle, C.R., Li, X. & Lytton, J. Novel role for K+-dependent Na+/Ca2+ exchangers in regulation of cytoplasmic free Ca2+ and contractility in arterial smooth muscle. Am. J. Physiol. Heart Circ. Physiol. 291, H1226–H1235 (2006).
Yamaji, M., Mahmoud, M., Evans, I.M. & Zachary, I.C. Neuropilin 1 is essential for gastrointestinal smooth muscle contractility and motility in aged mice. PLoS One 10, e0115563 (2015).
Lu, X. et al. Genome-wide association study in Han Chinese identifies four new susceptibility loci for coronary artery disease. Nat. Genet. 44, 890–894 (2012).
Hager, J. et al. Genome-wide association study in a Lebanese cohort confirms PHACTR1 as a major determinant of coronary artery stenosis. PLoS One 7, e38663 (2012).
The Coronary Artery Disease (C4D) Genetics Consortium. A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat. Genet. 43, 339–344 (2011).
O'Donnell, C.J. et al. Genome-wide association study for coronary artery calcification with follow-up in myocardial infarction. Circulation 124, 2855–2864 (2011).
Porcu, E. et al. A meta-analysis of thyroid-related traits reveals novel loci and gender-specific differences in the regulation of thyroid function. PLoS Genet. 9, e1003266 (2013).
Soler Artigas, M. et al. Genome-wide association and large-scale follow up identifies 16 new loci influencing lung function. Nat. Genet. 43, 1082–1090 (2011).
Lu, T. et al. REST and stress resistance in ageing and Alzheimer disease. Nature 507, 448–454 (2014).
Kar, R., Riquelme, M.A., Werner, S. & Jiang, J.X. Connexin 43 channels protect osteocytes against oxidative stress-induced cell death. J. Bone Miner. Res. 28, 1611–1621 (2013).
Dixit, D., Ghildiyal, R., Anto, N.P. & Sen, E. Chaetocin-induced ROS-mediated apoptosis involves ATM-YAP1 axis and JNK-dependent inhibition of glucose metabolism. Cell Death Dis. 5, e1212 (2014).
Chuikov, S., Levi, B.P., Smith, M.L. & Morrison, S.J. Prdm16 promotes stem cell maintenance in multiple tissues, partly by regulating oxidative stress. Nat. Cell Biol. 12, 999–1006 (2010).
Castellano, J. et al. Hypoxia stimulates low-density lipoprotein receptor-related protein-1 expression through hypoxia-inducible factor-1α in human vascular smooth muscle cells. Arterioscler. Thromb. Vasc. Biol. 31, 1411–1420 (2011).
Schlossmann, J. et al. Regulation of intracellular calcium by a signalling complex of IRAG, IP3 receptor and cGMP kinase Iβ. Nature 404, 197–201 (2000).
Nalls, M.A. et al. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson's disease. Nat. Genet. 46, 989–993 (2014).
Lambert, J.C. et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat. Genet. 45, 1452–1458 (2013).
Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427 (2014).
Wood, A.R. et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat. Genet. 46, 1173–1186 (2014).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Bulik-Sullivan, B.K. et al. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015).
Yang, J. et al. Genomic inflation factors under polygenic inheritance. Eur. J. Hum. Genet. 19, 807–812 (2011).
Magi, R., Lindgren, C.M. & Morris, A.P. Meta-analysis of sex-specific genome-wide association studies. Genet. Epidemiol. 34, 846–853 (2010).
Maller, J.B. et al. Bayesian refinement of association signals for 14 loci in 3 common diseases. Nat. Genet. 44, 1294–1301 (2012).
Nicolae, D.L. et al. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 6, e1000888 (2010).
Maurano, M.T. et al. Systematic localization of common disease-associated variation in regulatory DNA. Science 337, 1190–1195 (2012).
The GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 45, 580–585 (2013).
Pers, T.H. et al. Biological interpretation of genome-wide association studies using predicted gene functions. Nat. Commun. 6, 5890 (2015).
Chi, J.T. et al. Gene expression programs of human smooth muscle cells: tissue-specific differentiation and prognostic significance in breast cancers. PLoS Genet. 3, 1770–1784 (2007).
Bernstein, B.E. et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat. Biotechnol. 28, 1045–1048 (2010).
The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Winsvold, B.S. et al. Genetic analysis for a shared biological basis between migraine and coronary artery disease. Neurol. Genet. 1, e10 (2015).
Malik, R. et al. Shared genetic basis for migraine and ischemic stroke: a genome-wide analysis of common variants. Neurology 84, 2132–2145 (2015).
Ferrari, M.D., Klever, R.R., Terwindt, G.M., Ayata, C. & van den Maagdenberg, A.M.J.M. Migraine pathophysiology: lessons from mouse models and human genetics. Lancet Neurol. 14, 65–80 (2015).
Olesen, J., Burstein, R., Ashina, M. & Tfelt-Hansen, P. Origin of pain in migraine: evidence for peripheral sensitisation. Lancet Neurol. 8, 679–690 (2009).
Hadjikhani, N. et al. Mechanisms of migraine aura revealed by functional MRI in human visual cortex. Proc. Natl. Acad. Sci. USA 98, 4687–4692 (2001).
Lauritzen, M. Pathophysiology of the migraine aura. The spreading depression theory. Brain 117, 199–210 (1994).
Olesen, J. The role of nitric oxide (NO) in migraine, tension-type headache and cluster headache. Pharmacol. Ther. 120, 157–171 (2008).
Ashina, M., Hansen, J.M. & Olesen, J. Pearls and pitfalls in human pharmacological models of migraine: 30 years' experience. Cephalalgia 33, 540–553 (2013).
Read, S.J. & Parsons, A.A. Sumatriptan modifies cortical free radical release during cortical spreading depression: a novel antimigraine action for sumatriptan? Brain Res. 870, 44–53 (2000).
Anderson, C.A. et al. Data quality control in genetic case-control association studies. Nat. Protoc. 5, 1564–1573 (2010).
Winkler, T.W. et al. Quality control and conduct of genome-wide association meta-analyses. Nat. Protoc. 9, 1192–1212 (2014).
Delaneau, O., Marchini, J. & Zagury, J.-F. A linear complexity phasing method for thousands of genomes. Nat. Methods 9, 179–181 (2011).
Howie, B., Fuchsberger, C., Stephens, M., Marchini, J. & Abecasis, G.R. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet. 44, 955–959 (2012).
Browning, S.R. & Browning, B.L. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 81, 1084–1097 (2007).
Li, Y., Willer, C.J., Ding, J., Scheet, P. & Abecasis, G.R. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol. 34, 816–834 (2010).
Fuchsberger, C., Abecasis, G.R. & Hinds, D.A. minimac2: faster genotype imputation. Bioinformatics 31, 782–784 (2015).
The International HapMap 3 Consortium. Integrating common and rare genetic variation in diverse human populations. Nature 467, 52–58 (2010).
Schwarz, G. Estimating the dimension of a model. Ann. Stat. 6, 461–464 (1978).
Wright, F.A. et al. Heritability and genomics of gene expression in peripheral blood. Nat. Genet. 46, 430–437 (2014).
Richards, A.L. et al. Schizophrenia susceptibility alleles are enriched for alleles that affect gene expression in adult human brain. Mol. Psychiatry 17, 193–201 (2012).
Nyholt, D.R. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am. J. Hum. Genet. 74, 765–769 (2004).
Farh, K.K.-H. et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 518, 337–343 (2015).
Mi, H., Muruganujan, A., Casagrande, J.T. & Thomas, P.D. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 8, 1551–1566 (2013).
Fehrmann, R.S.N. et al. Gene expression analysis identifies global gene dosage sensitivity in cancer. Nat. Genet. 47, 115–125 (2015).
Frey, B.J. & Dueck, D. Clustering by passing messages between data points. Science 315, 972–976 (2007).
Acknowledgements
We thank the numerous individuals who contributed to sample collection, storage, handling, phenotyping, and genotyping for each of the individual cohorts. We also acknowledge the important contribution to research made by the study participants. We are grateful to H. Zhao (QIMR Berghofer Medical Research Institute) for helpful correspondence on the pathway analyses. We acknowledge the GTEx Consortium for support and contribution of pilot data. A list of study-specific acknowledgments and funding information can be found in the Supplementary Note.
Author information
Authors and Affiliations
Consortia
Contributions
P.G., V. Anttila, G.W.M., M.I.K., M. Kals, R. Mägi, K.P., E.H., E.L., A.G.U., L.C., E.M., L.M., A.-L.E., A.F.C., T.F.H., A.J.A., D.I.C., and D.R.N. performed the experiments. P.G., V. Anttila, B.S.W., P.P., T.E., T.H.P., K.-H.F., M. Muona, N.A.F., A.I., G.M., L.L., S.G.G., S. Steinberg, L.Q., H.H.H.A., D.A.H., J.-J.H., R. Malik, A.E.B., E.S., C.M.v.D., E.M., D.P.S., N.E., B.M.N., D.I.C., and D.R.N. performed the statistical analyses. P.G., V. Anttila, B.S.W., P.P., T.E., T.H.P., K.-H.F., E.C.-L., N.A.F., A.I., G.M., L.L., M. Kallela, T.M.F., S.G.G., S. Steinberg, M. Koiranen, L.Q., H.H.H.A., T.L., J.W., D.A.H., S.M.R., M.F., V. Artto, M. Kaunisto, S.V., R. Malik, M.I.K., M. Kals, R. Mägi, K.P., H.H., A.E.B., J.H., E.S., C.S., C.W., Z.C., K.H., E.L., L.M.P., A.-L.E., A.F.C., T.F.H., J.K., A.J.A., O.R., M.A.I., M.-R.J., D.P.S., M.W., G.D.S., N.E., M.J.D., B.M.N., J.O., D.I.C., D.R.N., and A.P. participated in data analysis and/or interpretation. P.G., V. Anttila, B.S.W., T.H.P., K.-H.F., E.C.-L., T.K., G.M.T., M. Kallela, C.R., A.H.S., G.B., M. Koiranen, T.L., M.S., M.G.H., M.F., V. Artto, M. Kaunisto, S.V., R. Malik, A.C.H., P.A.F.M., N.G.M., G.W.M., H.H., A.E.B., L.F., J.H., P.H.L., C.S., C.W., Z.C., B.M.-M., S. Schreiber, T.M., J.G.E., V.S., A.G.U., C.M.v.D., A.S., C.S.N., H.G., A.-L.E., A.F.C., T.F.H., T.W., A.J.A., O.R., M.-R.J., C.K., M.D.F., A.C.B., M.D., M.W., J.-A.Z., B.M.N., J.O., D.I.C., D.R.N., A.-P.S., J.E.B., P.M.R., and A.P. contributed materials and/or analysis tools. T.E., T.K., T.L., H.S., B.W.J.H.P., A.C.H., P.A.F.M., N.G.M., G.W.M., L.F., A.H., A.S., C.S.N., M. Männikkö, T.W., J.K., O.R., M.A.I., T.S., M.-R.J., A.M., C.K., D.P.S., M.D.F., A.M.J.M.v.d.M., J.-A.Z., D.I.B., G.D.S., K.S., N.E., B.M.N., J.O., D.I.C., D.R.N., and A.P. supervised the research. T.K., G.M.T., G.B., T.L., J.E.B., M.S., P.M.R., H.S., B.W.J.H.P., A.C.H., P.A.F.M., N.G.M., G.W.M., L.F., V.S., A.H., L.C., A.S., C.S.N., H.G., J.K., A.J.A., O.R., M.A.I., M.-R.J., A.M., C.K., D.P.S., M.D., A.M.J.M.v.d.M., D.I.B., G.D.S., N.E., M.J.D., B.M.N., D.I.C., D.R.N., and A.P. conceived and designed the study. P.G., V. Anttila, B.S.W., P.P., T.E., T.H.P., E.C.-L., H.H., B.M.N., J.O., D.I.C., D.R.N., and A.P. wrote the paper. All authors contributed to the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
T.W. has acted as a lecturer and consultant for H. Lundbeck A/S, Valby, Denmark. M.S. is a full-time employee of Bayer HealthCare, Germany.
Additional information
A full list of members and affiliations appears at the end of the paper and in the Supplementary Note.
Supplementary information
Supplementary Text and Figures
Supplementary Note, Supplementary Figures 3–15, and Supplementary Tables 1–25 (PDF 7376 kb)
Supplementary Figure 1
Forest plots of the 44 independently associated SNPs (PDF 2623 kb)
Supplementary Figure 2
Regional plots of the 44 independently associated SNPs (PDF 3517 kb)
Rights and permissions
About this article
Cite this article
Gormley, P., Anttila, V., Winsvold, B. et al. Meta-analysis of 375,000 individuals identifies 38 susceptibility loci for migraine. Nat Genet 48, 856–866 (2016). https://doi.org/10.1038/ng.3598
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3598
This article is cited by
-
Galcanezumab effects on incidence of headache after occurrence of triggers, premonitory symptoms, and aura in responders, non-responders, super-responders, and super non-responders
The Journal of Headache and Pain (2023)
-
Alterations in DNA methylation associate with reduced migraine and headache days after medication withdrawal treatment in chronic migraine patients: a longitudinal study
Clinical Epigenetics (2023)
-
Identifying causal genes for migraine by integrating the proteome and transcriptome
The Journal of Headache and Pain (2023)
-
Non-coding variants in VAMP2 and SNAP25 affect gene expression: potential implications in migraine susceptibility
The Journal of Headache and Pain (2023)
-
Genetics of migraine: where are we now?
The Journal of Headache and Pain (2023)