Abstract
We identified biallelic mutations in NANS, the gene encoding the synthase for N-acetylneuraminic acid (NeuNAc; sialic acid), in nine individuals with infantile-onset severe developmental delay and skeletal dysplasia. Patient body fluids showed an elevation in N-acetyl-D-mannosamine levels, and patient-derived fibroblasts had reduced NANS activity and were unable to incorporate sialic acid precursors into sialylated glycoproteins. Knockdown of nansa in zebrafish embryos resulted in abnormal skeletal development, and exogenously added sialic acid partially rescued the skeletal phenotype. Thus, NANS-mediated synthesis of sialic acid is required for early brain development and skeletal growth. Normal sialylation of plasma proteins was observed in spite of NANS deficiency. Exploration of endogenous synthesis, nutritional absorption, and rescue pathways for sialic acid in different tissues and developmental phases is warranted to design therapeutic strategies to counteract NANS deficiency and to shed light on sialic acid metabolism and its implications for human nutrition.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
Change history
06 March 2017
In the version of this article initially published, the name of author Torben Heise was given incorrectly as Thorben Heisse, and the name of author Valérie Cormier-Daire was given incorrectly as Valerie Cormier. The institutional affiliation for Delphine Heron was listed incorrectly as Institut IMAGINE, Hôpital Necker–Enfants Malades, Paris, France, and should have been listed as Département de Génétique Médicale et Centre de Référence Déficiences Intellectuelles, Groupe Hospitalier Pitié-Salpêtrière, Université Pierre et Marie Curie, Paris, France. The errors have been corrected in the HTML and PDF versions of the article.
References
Salvador-Carulla, L. et al. Intellectual developmental disorders: towards a new name, definition and framework for “mental retardation/intellectual disability” in ICD-11. World Psychiatry 10, 175–180 (2011).
American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders 5th edn (American Psychiatric Publishing, 2013).
de Ligt, J. et al. Diagnostic exome sequencing in persons with severe intellectual disability. N. Engl. J. Med. 367, 1921–1929 (2012).
Gilissen, C. et al. Genome sequencing identifies major causes of severe intellectual disability. Nature 511, 344–347 (2014).
van Karnebeek, C.D.M. & Stockler, S. Treatable inborn errors of metabolism causing intellectual disability: a systematic literature review. Mol. Genet. Metab. 105, 368–381 (2012).
Bonafé, L. et al. Nosology and classification of genetic skeletal disorders: 2015 revision. Am. J. Med. Genet. 167, 2869–2892 (2015).
Superti-Furga, A., Bonafé, L. & Rimoin, D.L. Molecular-pathogenetic classification of genetic disorders of the skeleton. Am. J. Med. Genet. 106, 282–293 (2001).
Camera, G., Camera, A., Di Rocco, M. & Gatti, R. Sponastrime dysplasia: report on two siblings with metal retardation. Pediatr. Radiol. 23, 611–614 (1993).
Geneviève, D. et al. Exclusion of the dymeclin and PAPSS2 genes in a novel form of spondyloepimetaphyseal dysplasia and mental retardation. Eur. J. Hum. Genet. 13, 541–546 (2005).
Tarailo-Graovac, M. et al. Exome sequencing and the management of neurometabolic disorders. N. Engl. J. Med. (in the press).
Cotton, T.R., Joseph, D.D., Jiao, W. & Parker, E.J. Probing the determinants of phosphorylated sugar-substrate binding for human sialic acid synthase. Biochim. Biophys. Acta 1844, 2257–2264 (2014).
Galuska, S.P. et al. Quantification of nucleotide-activated sialic acids by a combination of reduction and fluorescent labeling. Anal. Chem. 82, 4591–4598 (2010).
Büll, C. et al. Sialic acid glycoengineering using an unnatural sialic acid for the detection of sialoglycan biosynthesis defects and on-cell synthesis of siglec ligands. ACS Chem. Biol. 10, 2353–2363 (2015).
Riemersma, M. et al. Disease mutations in CMP–sialic acid transporter SLC35A1 result in abnormal α-dystroglycan O-mannosylation, independent from sialic acid. Hum. Mol. Genet. 24, 2241–2246 (2015).
Link, V., Shevchenko, A. & Heisenberg, C.P. Proteomics of early zebrafish embryos. BMC Dev. Biol. 6, 1 (2006).
Eisen, J.S. & Smith, J.C. Controlling morpholino experiments: don't stop making antisense. Development 135, 1735–1743 (2008).
Javidan, Y. & Schilling, T.F. Development of cartilage and bone. Methods Cell Biol. 76, 415–436 (2004).
Wang, B. & Brand-Miller, J. The role and potential of sialic acid in human nutrition. Eur. J. Clin. Nutr. 57, 1351–1369 (2003).
Simpson, M.A. et al. Infantile-onset symptomatic epilepsy syndrome caused by a homozygous loss-of-function mutation of GM3 synthase. Nat. Genet. 36, 1225–1229 (2004).
Hu, H. et al. ST3GAL3 mutations impair the development of higher cognitive functions. Am. J. Hum. Genet. 89, 407–414 (2011).
Fragaki, K. et al. Refractory epilepsy and mitochondrial dysfunction due to GM3 synthase deficiency. Eur. J. Hum. Genet. 21, 528–534 (2013).
Boccuto, L. et al. A mutation in a ganglioside biosynthetic enzyme, ST3GAL5, results in salt & pepper syndrome, a neurocutaneous disorder with altered glycolipid and glycoprotein glycosylation. Hum. Mol. Genet. 23, 418–433 (2014).
Mohamed, M. et al. Intellectual disability and bleeding diathesis due to deficient CMP–sialic acid transport. Neurology 81, 681–687 (2013).
Roughley, P.J., White, R.J. & Santer, V. Comparison of proteoglycans extracted from high and low weight–bearing human articular cartilage, with particular reference to sialic acid content. J. Biol. Chem. 256, 12699–12704 (1981).
Vincent, K. & Durrant, M.C. A structural and functional model for human bone sialoprotein. J. Mol. Graph. Model. 39, 108–117 (2013).
Sodek, J., Ganss, B. & McKee, M.D. Osteopontin. Crit. Rev. Oral Biol. Med. 11, 279–303 (2000).
Wang, B. Sialic acid is an essential nutrient for brain development and cognition. Annu. Rev. Nutr. 29, 177–222 (2009).
Wang, B. Molecular mechanism underlying sialic acid as an essential nutrient for brain development and cognition. Adv. Nutr. 3, 465S–472S (2012).
Sprenger, N. & Duncan, P.I. Sialic acid utilization. Adv. Nutr. 3, 392S–397S (2012).
Salama, I. et al. No overall hyposialylation in hereditary inclusion body myopathy myoblasts carrying the homozygous M712T GNE mutation. Biochem. Biophys. Res. Commun. 328, 221–226 (2005).
Malicdan, M.C., Noguchi, S., Hayashi, Y.K., Nonaka, I. & Nishino, I. Prophylactic treatment with sialic acid metabolites precludes the development of the myopathic phenotype in the DMRV-hIBM mouse model. Nat. Med. 15, 690–695 (2009).
Oetke, C. et al. Evidence for efficient uptake and incorporation of sialic acid by eukaryotic cells. Eur. J. Biochem 268, 4553–4561 (2001).
Nöhle, U. & Schauer, R. Uptake, metabolism and excretion of orally and intravenously administered, 14C- and 3H-labeled N-acetylneuraminic acid mixture in the mouse and rat. Hoppe-Seyler's Z. Physiol. Chem. 362, 1495–1506 (1981).
Tangvoranuntakul, P. et al. Human uptake and incorporation of an immunogenic nonhuman dietary sialic acid. Proc. Natl. Acad. Sci. USA 100, 12045–12050 (2003).
Samraj, A.N. et al. A red meat–derived glycan promotes inflammation and cancer progression. Proc. Natl. Acad. Sci. USA 112, 542–547 (2015).
Bode, L. Human milk oligosaccharides: every baby needs a sugar mama. Glycobiology 22, 1147–1162 (2012).
Fuhrer, A. et al. Milk sialyllactose influences colitis in mice through selective intestinal bacterial colonization. J. Exp. Med. 207, 2843–2854 (2010).
Röhrig, C.H., Choi, S.S. & Baldwin, N. The nutritional role of free sialic acid, a human milk monosaccharide, and its application as a functional food ingredient. Crit. Rev. Food Sci. Nutr. (doi:10.1080/10408398.2015.1040113 (2016)).
Van der Auwera, G.A. et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinformatics 43, 1–33 (2013).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010).
Yang, J. et al. The I-TASSER Suite: protein structure and function prediction. Nat. Methods 12, 7–8 (2015).
Gunawan, J. et al. Structural and mechanistic analysis of sialic acid synthase NeuB from Neisseria meningitidis in complex with Mn2+, phosphoenolpyruvate, and N-acetylmannosaminitol. J. Biol. Chem. 280, 3555–3563 (2005).
Hamada, T. et al. Solution structure of the antifreeze-like domain of human sialic acid synthase. Protein Sci. 15, 1010–1016 (2006).
Rajavel, K.S. & Neufeld, E.F. Nonsense-mediated decay of human HEXA mRNA. Mol. Cell. Biol. 21, 5512–5519 (2001).
Wishart, D.S. et al. HMDB 3.0—the Human Metabolome Database in 2013. Nucleic Acids Res. 41, D801–D807 (2013).
Engelke, U.F. et al. NMR spectroscopic studies on the late onset form of 3-methylglutaconic aciduria type I and other defects in leucine metabolism. NMR Biomed. 19, 271–278 (2006).
Valianpour, F., Abeling, N.G., Duran, M., Huijmans, J.G. & Kulik, W. Quantification of free sialic acid in urine by HPLC–electrospray tandem mass spectrometry: a tool for the diagnosis of sialic acid storage disease. Clin. Chem. 50, 403–409 (2004).
van der Ham, M. et al. Liquid chromatography–tandem mass spectrometry assay for the quantification of free and total sialic acid in human cerebrospinal fluid. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 878, 1098–1102 (2010).
Jansen, J.C. et al. CCDC115 deficiency causes a disorder of Golgi homeostasis with abnormal protein glycosylation. Am. J. Hum. Genet. 98, 310–321 (2016).
van Scherpenzeel, M., Steenbergen, G., Morava, E., Wevers, R.A. & Lefeber, D.J. High-resolution mass spectrometry glycoprofiling of intact transferrin for diagnosis and subtype identification in the congenital disorders of glycosylation. Transl. Res. 166, 639–649 (2015).
Acknowledgements
We thank M. Filocamo at the Gaslini Biobank (Genoa, Italy) for a fibroblast line for patient 1. We thank A. Reymond (CIG, FBM, Université de Lausanne) and his laboratory for lymphocyte immortalization. We thank C. Chiesa for Sanger sequencing and sample handling and shipment; S. de Boer for excellent technical assistance; B. Toh at the University of British Columbia for metabolic sample handling; X. Han for Sanger sequencing; B. Sayson for consenting and data management; M. Higginson for DNA extraction and sample handling; and A. Ghani for administrative assistance. We also thank R. Houben for skillfully preparing Figure 4 and A. Bandi for all other figures. We are grateful to our clinical colleagues in Dolo, Genoa, Lausanne, Manchester, Paris, Reggio Emilia, Tokyo, Treviso, and Vancouver for patient management. A.S.-F. dedicates this paper to the memory of Paolo Durand who pointed out the relationship between sialic acid metabolism and IDD to him in 1980. Finally, we wish to thank the patients reported here as well as their parents for the enthusiasm they showed for our research efforts, for their patience, which was challenged by the studies lasting many years, and for their repeated donation of biological samples. They have been the source of continuous motivation for us.
This work has been supported by funding from the Leenaards Foundation in Lausanne, Switzerland; the Faculty of Biology and Medicine of the University of Lausanne; the BC Children's Hospital Foundation (Treatable Intellectual Disability Endeavour in British Columbia: First Collaborative Area of Innovation); Genome BC (grant SOF-195); the Rare Diseases Foundation; the Rare Diseases Models and Mechanisms Network; the Canadian Institutes of Health Research (grant 301221); and the Dutch Organization for Scientific Research, ZONMW (Medium Investment Grant 40-00506-98-9001 and VIDI Grant 91713359 to D.J.L.). The zebrafish studies were supported by funding to X.-Y.W. from the Canadian Rare Disease Models and Mechanisms Network, the Brain Canada Foundation, the Natural Sciences and Engineering Research Council of Canada (NSERC), and the Canada Foundation for Innovation (CFI). The informatics infrastructures were supported by Genome BC and Genome Canada (ABC4DE Project) as well as by the Vital-IT Project of the Swiss Institute of Bioinformatics (SIB; Lausanne, Switzerland). C.J.R. is funded by a Canadian Institutes of Health Research New Investigator Award. C.D.M.v.K. is a recipient of the Michael Smith Foundation for Health Research Scholar Award (Vancouver, Canada). E.G. is supported by a Marie Skłodowska-Curie fellowship (MSCA-IF-661491).
Author information
Authors and Affiliations
Contributions
C.D.M.v.K., L.B., S. Unger, R.A.W., and A.S.-F. conceived the study and coordinated and supervised the different teams. A.S.-F., L.B., C.D.M.v.K., T.D., A.C., M.I.V.A., C.J.R., J.H., L.G., L.T., V.C.-D., D.H., D.D., C.B., I.M., and S. Uchikawa recruited the patients, reviewed the clinical and radiographic features, and obtained biological materials from patients. A.S.-F., S. Unger, and G.N. reviewed the radiographic data. J.R. performed the bone marrow studies. Andrea Rossi reviewed the cerebral imaging. K. Harshman, B.J.S., B.C.-X., S.B., B.R.-B., H.R., C.R., M.T.-G., W.W.W., and A.d.B. were responsible for exome sequencing, haplotype reconstruction, Sanger sequencing, database studies, and mRNA–cDNA studies. R.A.W., L.A.J.K., E.v.d.H., and U.F.E. performed the metabolomics studies. Antonio Rossi studied ManNAc incorporation in fibroblasts. T. Hennet performed the lectin binding studies. A.A., K. Huijben, F.Z., and D.J.L. performed the NANS enzyme assays. D.J.L., A.A., T. Heisse, and T.B. studied the incorporation of sialic acid precursors in lymphocytes and fibroblasts. E.G. and G.S.-F. obtained the NANS three-dimensional model and mapped the affected amino acid residues. X.-Y.W. and K.B.-A. generated and phenotyped the zebrafish model. C.D.M.v.K. and A.S.-F. prepared the manuscript with contributions from all co-authors. All co-authors edited and reviewed the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
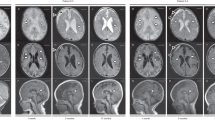
Supplementary Figure 1 Brain magnetic resonance imaging in patient 9 at age 3.6 years.
(a–d) Axial T2-weighted images show a marked degree of ventriculomegaly. Note the diencephalic–mesencephalic junction dysplasia, characterized by a lack of separation between the hypothalamus and midbrain (arrowheads in a). In the nucleobasal region, bilaterally, there are dilated perivascular spaces (arrowheads in b) and an abnormal morphology of the basal nuclei, with prominent thalami (labeled T in c) and maloriented putamen and caudate heads (labeled P and C, respectively, in c). Note also the bilateral abnormal orientation of the posterior arms of the sylvian fissure (empty arrows in d). (e) An axial T1-weighted image obtained with a volumetric technique shows thickening of the left perisylvian cortex, consistent with polymicrogyria. Cortical abnormality is less prominent controlaterally, despite the abnormal orientation of the sylvian fissure. (f) A left parasagittal T1-weighted image shows abnormal orientation of the sylvian fissure (arrows), which is continuous with the sulci of the convexity without a normal posterior delimitation; this pattern, which was also visible controlaterally (data not shown), is typical of perisylvian polymicrogyria. (g) A midsagittal T1-weighted image shows shortened, hypoplastic corpus callosum with a thin splenium (thin arrows) and lacking a proper rostrum (thick arrow); also note the absence of a well-defined anterior commissure (arrowhead).
Supplementary Figure 2 Capillary electrophoresis analysis of NANS cDNA retrotranscribed and amplified from RNA extracted from cell cultures.
(a) Schematic of the exon–intron structure of the NANS gene. Asterisks indicate mutations with a putative effect on exon splicing (see Table 1 for details): EXins: c.385_386insT, exonic single-nucleotide insertion producing a frameshift and creating a premature termination codon; SPLko: c.488+1G>T, a canonical splice donor site mutation; INTindel: c.449–10delGATTACinsATGG, an intronic indel 5′ of exon 4. (b) Pedigrees of patients 1, 3, and 4, and corresponding capillary electrophoresis traces of semiquantitative RT-PCRs spanning all analyzed NANS mutations. We assessed unprocessed PCR products obtained with the same primer pairs and allowing the simultaneous ampli-fication of the wild-type and the mutant mRNA (cDNA) forms, indicated on the right, within the same reaction. See the main text for discussion of the results.
Supplementary Figure 3 Expression of NANS in human brain as extracted from the Brainspan database.
In brain, NANS is expressed at significantly higher levels in the thalamus than in other brain regions (top) (http://developinghumanbrain.org/). Expression seems to be highest in the dorsal thalamus (bottom), which is an important relay station for the limbic circuit (The Human Nervous System 439–468 Academic Press, 1990) and participates in learning and memory processes (J. Neurosci. 34, 15340–15346, 2014), which might contribute to the developmental delay in patients with NANS deficiency.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–3, Supplementary Tables 1–5 and Supplementary Note. (PDF 2775 kb)
Rights and permissions
About this article
Cite this article
van Karnebeek, C., Bonafé, L., Wen, XY. et al. NANS-mediated synthesis of sialic acid is required for brain and skeletal development. Nat Genet 48, 777–784 (2016). https://doi.org/10.1038/ng.3578
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3578
This article is cited by
-
Crude enzyme immobilization-based cell-free system for efficient N-acetylneuraminic acid biosynthesis aided by N-terminal coding sequence screening
Systems Microbiology and Biomanufacturing (2023)
-
Clinical and molecular findings in three Japanese patients with N-acetylneuraminic acid synthetase-congenital disorder of glycosylation (NANS-CDG)
Scientific Reports (2022)
-
Switching azide and alkyne tags on bioorthogonal reporters in metabolic labeling of sialylated glycoconjugates: a comparative study
Scientific Reports (2022)
-
Neonatal encephalopathy plasma metabolites are associated with neurodevelopmental outcomes
Pediatric Research (2022)
-
Treatable inherited metabolic disorders causing intellectual disability: 2021 review and digital app
Orphanet Journal of Rare Diseases (2021)