Abstract
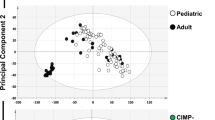
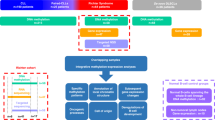
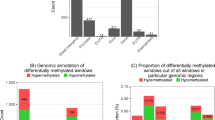
Although Burkitt lymphomas and follicular lymphomas both have features of germinal center B cells, they are biologically and clinically quite distinct. Here we performed whole-genome bisulfite, genome and transcriptome sequencing in 13 IG-MYC translocation–positive Burkitt lymphoma, nine BCL2 translocation–positive follicular lymphoma and four normal germinal center B cell samples. Comparison of Burkitt and follicular lymphoma samples showed differential methylation of intragenic regions that strongly correlated with expression of associated genes, for example, genes active in germinal center dark-zone and light-zone B cells. Integrative pathway analyses of regions differentially methylated in Burkitt and follicular lymphomas implicated DNA methylation as cooperating with somatic mutation of sphingosine phosphate signaling, as well as the TCF3-ID3 and SWI/SNF complexes, in a large fraction of Burkitt lymphomas. Taken together, our results demonstrate a tight connection between somatic mutation, DNA methylation and transcriptional control in key B cell pathways deregulated differentially in Burkitt lymphoma and other germinal center B cell lymphomas.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Lai, A.Y. et al. DNA methylation profiling in human B cells reveals immune regulatory elements and epigenetic plasticity at Alu elements during B-cell activation. Genome Res. 23, 2030–2041 (2013).
Lee, S.T. et al. A global DNA methylation and gene expression analysis of early human B-cell development reveals a demethylation signature and transcription factor network. Nucleic Acids Res. 40, 11339–11351 (2012).
Shaknovich, R. et al. DNA methyltransferase 1 and DNA methylation patterning contribute to germinal center B-cell differentiation. Blood 118, 3559–3569 (2011).
Basso, K. & Dalla-Favera, R. Germinal centres and B cell lymphomagenesis. Nat. Rev. Immunol. 15, 172–184 (2015).
Lenz, G. & Staudt, L.M. Aggressive lymphomas. N. Engl. J. Med. 362, 1417–1429 (2010).
Küppers, R. & Dalla-Favera, R. Mechanisms of chromosomal translocations in B cell lymphomas. Oncogene 20, 5580–5594 (2001).
Dave, S.S. et al. Molecular diagnosis of Burkitt's lymphoma. N. Engl. J. Med. 354, 2431–2442 (2006).
Richter, J. et al. Recurrent mutation of the ID3 gene in Burkitt lymphoma identified by integrated genome, exome and transcriptome sequencing. Nat. Genet. 44, 1316–1320 (2012).
Schmitz, R. et al. Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature 490, 116–120 (2012).
Loeffler, M. et al. Genomic and epigenomic co-evolution in follicular lymphomas. Leukemia 29, 456–463 (2015).
Morin, R.D. et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat. Genet. 42, 181–185 (2010).
Morin, R.D. et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature 476, 298–303 (2011).
Okosun, J. et al. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat. Genet. 46, 176–181 (2014).
Pasqualucci, L. et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature 471, 189–195 (2011).
Victora, G.D. et al. Identification of human germinal center light and dark zone cells and their relationship to human B-cell lymphomas. Blood 120, 2240–2248 (2012).
Otto, C., Stadler, P.F. & Hoffmann, S. Fast and sensitive mapping of bisulfite-treated sequencing data. Bioinformatics 28, 1698–1704 (2012).
Ernst, J. et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473, 43–49 (2011).
Hahn, M.A. et al. Loss of the Polycomb mark from bivalent promoters leads to activation of cancer-promoting genes in colorectal tumors. Cancer Res. 74, 3617–3629 (2014).
Hovestadt, V. et al. Decoding the regulatory landscape of medulloblastoma using DNA methylation sequencing. Nature 510, 537–541 (2014).
Weidensdorfer, D. et al. Control of c-myc mRNA stability by IGF2BP1-associated cytoplasmic RNPs. RNA 15, 104–115 (2009).
Hummel, M. et al. A biologic definition of Burkitt's lymphoma from transcriptional and genomic profiling. N. Engl. J. Med. 354, 2419–2430 (2006).
Love, C. et al. The genetic landscape of mutations in Burkitt lymphoma. Nat. Genet. 44, 1321–1325 (2012).
Muppidi, J.R. et al. Loss of signalling via Galpha13 in germinal centre B-cell–derived lymphoma. Nature 516, 254–258 (2014).
Rohde, M. et al. Recurrent RHOA mutations in pediatric Burkitt lymphoma treated according to the NHL-BFM protocols. Genes Chromosom. Cancer 53, 911–916 (2014).
O'Hayre, M. et al. The emerging mutational landscape of G proteins and G-protein–coupled receptors in cancer. Nat. Rev. Cancer 13, 412–424 (2013).
Takuwa, N. et al. Tumor-suppressive sphingosine-1-phosphate receptor-2 counteracting tumor-promoting sphingosine-1-phosphate receptor-1 and sphingosine kinase 1—Jekyll hidden behind Hyde. Am. J. Cancer Res. 1, 460–481 (2011).
Morin, R.D. et al. Mutational and structural analysis of diffuse large B-cell lymphoma using whole-genome sequencing. Blood 122, 1256–1265 (2013).
Abraham, B.J., Cui, K., Tang, Q. & Zhao, K. Dynamic regulation of epigenomic landscapes during hematopoiesis. BMC Genomics 14, 193 (2013).
Scott, C.L. et al. Role of the chromobox protein CBX7 in lymphomagenesis. Proc. Natl. Acad. Sci. USA 104, 5389–5394 (2007).
Dykhuizen, E.C. et al. BAF complexes facilitate decatenation of DNA by topoisomerase IIα. Nature 497, 624–627 (2013).
Klapper, W. et al. Patient age at diagnosis is associated with the molecular characteristics of diffuse large B-cell lymphoma. Blood 119, 1882–1887 (2012).
Hasselblatt, M. et al. Nonsense mutation and inactivation of SMARCA4 (BRG1) in an atypical teratoid/rhabdoid tumor showing retained SMARCB1 (INI1) expression. Am. J. Surg. Pathol. 35, 933–935 (2011).
Hasselblatt, M. et al. SMARCA4-mutated atypical teratoid/rhabdoid tumors are associated with inherited germline alterations and poor prognosis. Acta Neuropathol. 128, 453–456 (2014).
Schneppenheim, R. et al. Germline nonsense mutation and somatic inactivation of SMARCA4/BRG1 in a family with rhabdoid tumor predisposition syndrome. Am. J. Hum. Genet. 86, 279–284 (2010).
Witkowski, L. et al. Germline and somatic SMARCA4 mutations characterize small cell carcinoma of the ovary, hypercalcemic type. Nat. Genet. 46, 438–443 (2014).
Betts, M.J. et al. Mechismo: predicting the mechanistic impact of mutations and modifications on molecular interactions. Nucleic Acids Res. 43, e10 (2015).
Kulis, M. et al. Whole-genome fingerprint of the DNA methylome during human B cell differentiation. Nat. Genet. 47, 746–756 (2015).
Lister, R. et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature 471, 68–73 (2011).
Bibikova, M. et al. High density DNA methylation array with single CpG site resolution. Genomics 98, 288–295 (2011).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Jones, D.T. et al. Recurrent somatic alterations of FGFR1 and NTRK2 in pilocytic astrocytoma. Nat. Genet. 45, 927–932 (2013).
Rimmer, A. et al. Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nat. Genet. 46, 912–918 (2014).
Hoffmann, S. et al. Fast mapping of short sequences with mismatches, insertions and deletions using index structures. PLoS Comput. Biol. 5, e1000502 (2009).
Zhang, N.R. & David, O.S. Model selection for high-dimensional, multisequence change-point problems. Stat. Sin. 22, 1507 (2012).
Anders, S. & Huber, W. Differential expression analysis for sequence count data. Genome Biol. 11, R106 (2010).
Karolchik, D. et al. The UCSC Genome Browser database: 2014 update. Nucleic Acids Res. 42, D764–D770 (2014).
ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Flicek, P. et al. Ensembl 2014. Nucleic Acids Res. 42, D749–D755 (2014).
Ernst, J. & Kellis, M. ChromHMM: automating chromatin-state discovery and characterization. Nat. Methods 9, 215–216 (2012).
Dürr, H. et al. X-ray structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with DNA. Cell 121, 363–373 (2005).
Eswar, N. et al. Comparative protein structure modeling using Modeller. Curr. Protoc. Bioinformatics 47, 5.6.1–5.6.32 (2006).
Sharma, V. et al. Crystal structure of Mycobacterium tuberculosis SecA, a preprotein translocating ATPase. Proc. Natl. Acad. Sci. USA 100, 2243–2248 (2003).
Huber, W. et al. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 18, S96–S104 (2002).
Bentink, S. et al. Pathway activation patterns in diffuse large B-cell lymphomas. Leukemia 22, 1746–1754 (2008).
Wright, G. et al. A gene expression–based method to diagnose clinically distinct subgroups of diffuse large B cell lymphoma. Proc. Natl. Acad. Sci. USA 100, 9991–9996 (2003).
Acknowledgements
This study has been supported by the German Ministry of Science and Education (BMBF) in the framework of the ICGC MMML-Seq project (01KU1002A-J) and the MMML-MYC-SYS project (036166B), the European Union in the framework of the BLUEPRINT Project (HEALTH-F5-2011-282510) and the KinderKrebsInitiative Buchholz/Holm-Seppensen and LIFE (Leipzig Research Center for Civilization Diseases), Leipzig University. LIFE is funded by the European Union, the European Regional Development Fund (ERDF), the European Social Fund (ESF) and the Free State of Saxony. WGBS was additionally supported by NGFNplus (BMBF, 01GS0883) and the DKFZ–Heidelberg Center for Personalized Oncology (DKFZ-HIPO). Former grant support of MMML by the Deutsche Krebshilfe (2003–2011) is gratefully acknowledged. J.R. is supported by the Dr. Werner Jackstädt Foundation in the framework of a Junior Excellence Research Group on 'Mechanisms of B-Cell Lymphomagenesis in the Senium as Basic Principle for the Development of Age-Adjusted Therapy Regimes' (S134-10.100).
Author information
Authors and Affiliations
Consortia
Contributions
A.H., O.A., R.K., P.L., R.S., S. Hoffmann and B.R. conceived and designed the experiments. W.W., A.H., A.K.B., J.G., J.R., J.K., R.W., S.E., H.H.D.K., W.K., D.L., C. López, S.P., I.V., P.R., M. Schilhabel, M. Szczepanowski, L.T. and R.K. performed the experiments. H.K., S.H.B., M.J.B., B.H., R.E., P.F., C. Lawerenz, J.H.A.M., M. Schlesner, P.F.S., H.G.S. and S. Hoffmann performed statistical analysis. H.K., S.H.B., G.D., V.H., D.R., F.J., C.O., M.H., M. Kreuz, M. Kulis, I.N., M. Rosolowski, R.B.R., M. Schlesner, S. Hoffmann and B.R. analyzed the data. H.K., M.A.W., M.J.B., E.C.-d.-S.-P., G.D., B.H., F.J., Q.L., C.O., J.A., B.B., A.C., H.G.D., S.E., R.E., P.F., S. Haas, D.K., H.H.D.K., W.K., M. Kreuz, C. Lawerenz, D.L., M.L., R.A.F.M., J.H.A.M., J.I.M.-S., P.M., M. Rohde, P.R., M. Schilhabel, M. Schlesner, L.T., H.G.S. and S. Hoffmann contributed reagents, materials and/or analysis tools. H.K., S.H.B., W.W., A.H., P.L., R.S., S. Hoffmann and B.R. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
A full list of members and affiliations appears in the Supplementary Note.
A full list of members and affiliations appears in the Supplementary Note.
Supplementary information
Supplementary Text and Figures
Supplementary Note, Supplementary Figures 1–23 and Supplementary Tables 1–4, 6, 9–11, 13, 14 and 16–18. (PDF 4797 kb)
Supplementary Table 5
Statistics of whole-genome sequencing. (XLSX 15 kb)
Supplementary Table 7
Annotation of DMRs. (XLSX 7648 kb)
Supplementary Table 8
Correlating DMRs. (XLSX 990 kb)
Supplementary Table 12
Statistics of transcriptome arrays. (XLSX 29 kb)
Supplementary Table 15
SWI/SNF SNVs. (XLSX 14 kb)
Rights and permissions
About this article
Cite this article
Kretzmer, H., Bernhart, S., Wang, W. et al. DNA methylome analysis in Burkitt and follicular lymphomas identifies differentially methylated regions linked to somatic mutation and transcriptional control. Nat Genet 47, 1316–1325 (2015). https://doi.org/10.1038/ng.3413
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3413
This article is cited by
-
Molecular characterization of Richter syndrome identifies de novo diffuse large B-cell lymphomas with poor prognosis
Nature Communications (2023)
-
Mosaic chromosomal alterations in peripheral blood leukocytes of children in sub-Saharan Africa
Nature Communications (2023)
-
Genome-wide DNA methylation reveals potential epigenetic mechanism of age-dependent viral susceptibility in grass carp
Immunity & Ageing (2022)
-
Burkitt lymphoma
Nature Reviews Disease Primers (2022)
-
Research progress on epigenetics of small B-cell lymphoma
Clinical and Translational Oncology (2022)