Abstract
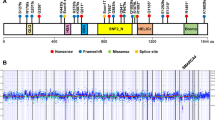
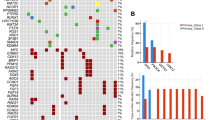
While investigating cohorts of unclassified sarcomas by RNA sequencing, we identified 19 cases with inactivation of SMARCA4, which encodes an ATPase subunit of BAF chromatin-remodeling complexes1,2. Clinically, the cases were all strikingly similar, presenting as compressive mediastino-pulmonary masses in 30- to 35-year-old adults with a median survival time of 7 months. To help define the nosological relationships of these tumors, we compared their transcriptomic profiles with those of SMARCA4-mutated small-cell carcinomas of the ovary, hypercalcemic type (SCCOHTs)3,4,5, SMARCB1-inactivated malignant rhabdoid tumors6 (MRTs) and lung carcinomas (of which 10% display SMARCA4 mutations7). Gene profiling analyses demonstrated that these tumors were distinct from lung carcinomas but related to MRTs and SCCOHTs. Transcriptome analyses, further validated by immunohistochemistry, highlighted strong expression of SOX2, a marker that supports the differential diagnosis of these tumors from SMARCA4-deficient lung carcinomas. The prospective recruitment of cases confirmed this new category of 'SMARCA4-deficient thoracic sarcomas' as readily recognizable in clinical practice, providing opportunities to tailor their therapeutic management.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Wang, W. et al. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 10, 2117–2130 (1996).
Khavari, P.A., Peterson, C.L., Tamkun, J.W., Mendel, D.B. & Crabtree, G.R. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature 366, 170–174 (1993).
Witkowski, L. et al. Germline and somatic SMARCA4 mutations characterize small cell carcinoma of the ovary, hypercalcemic type. Nat. Genet. 46, 438–443 (2014).
Jelinic, P. et al. Recurrent SMARCA4 mutations in small cell carcinoma of the ovary. Nat. Genet. 46, 424–426 (2014).
Ramos, P. et al. Small cell carcinoma of the ovary, hypercalcemic type, displays frequent inactivating germline and somatic mutations in SMARCA4. Nat. Genet. 46, 427–429 (2014).
Versteege, I. et al. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature 394, 203–206 (1998).
Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550 (2014).
Dykhuizen, E.C. et al. BAF complexes facilitate decatenation of DNA by topoisomerase IIα. Nature 497, 624–627 (2013).
Hargreaves, D.C. & Crabtree, G.R. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 21, 396–420 (2011).
Kadoch, C. et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat. Genet. 45, 592–601 (2013).
Klochendler-Yeivin, A. et al. The murine SNF5/INI1 chromatin remodeling factor is essential for embryonic development and tumor suppression. EMBO Rep. 1, 500–506 (2000).
Roberts, C.W., Leroux, M.M., Fleming, M.D. & Orkin, S.H. Highly penetrant, rapid tumorigenesis through conditional inversion of the tumor suppressor gene Snf5. Cancer Cell 2, 415–425 (2002).
Bultman, S.J. et al. Characterization of mammary tumors from Brg1 heterozygous mice. Oncogene 27, 460–468 (2008).
Witkowski, L. et al. Familial rhabdoid tumour 'avant la lettre'—from pathology review to exome sequencing and back again. J. Pathol. 231, 35–43 (2013).
Hasselblatt, M. et al. Nonsense mutation and inactivation of SMARCA4 (BRG1) in an atypical teratoid/rhabdoid tumor showing retained SMARCB1 (INI1) expression. Am. J. Surg. Pathol. 35, 933–935 (2011).
Guillou, L., Wadden, C., Coindre, J.M., Krausz, T. & Fletcher, C.D. “Proximal-type” epithelioid sarcoma, a distinctive aggressive neoplasm showing rhabdoid features. Clinicopathologic, immunohistochemical, and ultrastructural study of a series. Am. J. Surg. Pathol. 21, 130–146 (1997).
Varela, I. et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature 469, 539–542 (2011).
French, C.A. et al. BRD4 bromodomain gene rearrangement in aggressive carcinoma with translocation t(15;19). Am. J. Pathol. 159, 1987–1992 (2001).
Modena, P. et al. SMARCB1/INI1 tumor suppressor gene is frequently inactivated in epithelioid sarcomas. Cancer Res. 65, 4012–4019 (2005).
Le Loarer, F. et al. Consistent SMARCB1 homozygous deletions in epithelioid sarcoma and in a subset of myoepithelial carcinomas can be reliably detected by FISH in archival material. Genes Chromosom. Cancer 53, 475–486 (2014).
Sullivan, L.M., Folpe, A.L., Pawel, B.R., Judkins, A.R. & Biegel, J.A. Epithelioid sarcoma is associated with a high percentage of SMARCB1 deletions. Mod. Pathol. 26, 385–392 (2013).
Imielinski, M. et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 150, 1107–1120 (2012).
Seo, J.S. et al. The transcriptional landscape and mutational profile of lung adenocarcinoma. Genome Res. 22, 2109–2119 (2012).
Matsubara, D. et al. Lung cancer with loss of BRG1/BRM, shows epithelial mesenchymal transition phenotype and distinct histologic and genetic features. Cancer Sci. 104, 266–273 (2013).
Fukuoka, J. et al. Chromatin remodeling factors and BRM/BRG1 expression as prognostic indicators in non–small cell lung cancer. Clin. Cancer Res. 10, 4314–4324 (2004).
Orvis, T. et al. BRG1/SMARCA4 inactivation promotes non–small cell lung cancer aggressiveness by altering chromatin organization. Cancer Res. 74, 6486–6498 (2014).
Calderaro, J. et al. SMARCB1/INI1 inactivation in renal medullary carcinoma. Histopathology 61, 428–435 (2012).
Torchia, J. et al. Molecular subgroups of atypical teratoid rhabdoid tumours in children: an integrated genomic and clinicopathological analysis. Lancet Oncol. 16, 569–582 (2015).
Boyer, L.A. et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122, 947–956 (2005).
Ben-Porath, I. et al. An embryonic stem cell–like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet. 40, 499–507 (2008).
Leis, O. et al. Sox2 expression in breast tumours and activation in breast cancer stem cells. Oncogene 31, 1354–1365 (2012).
Kadoch, C. & Crabtree, G.R. Reversible disruption of mSWI/SNF (BAF) complexes by the SS18-SSX oncogenic fusion in synovial sarcoma. Cell 153, 71–85 (2013).
Ferri, A.L. et al. Sox2 deficiency causes neurodegeneration and impaired neurogenesis in the adult mouse brain. Development 131, 3805–3819 (2004).
Wang, W. et al. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 15, 5370–5382 (1996).
Doan, D.N. et al. Loss of the INI1 tumor suppressor does not impair the expression of multiple BRG1-dependent genes or the assembly of SWI/SNF enzymes. Oncogene 23, 3462–3473 (2004).
Wilson, B.G. et al. Residual complexes containing SMARCA2 (BRM) underlie the oncogenic drive of SMARCA4 (BRG1) mutation. Mol. Cell. Biol. 34, 1136–1144 (2014).
Wang, X. et al. Oncogenesis caused by loss of the SNF5 tumor suppressor is dependent on activity of BRG1, the ATPase of the SWI/SNF chromatin remodeling complex. Cancer Res. 69, 8094–8101 (2009).
Kahali, B. et al. The silencing of the SWI/SNF subunit and anticancer gene BRM in Rhabdoid tumors. Oncotarget 5, 3316–3332 (2014).
Hoffman, G.R. et al. Functional epigenetics approach identifies BRM/SMARCA2 as a critical synthetic lethal target in BRG1-deficient cancers. Proc. Natl. Acad. Sci. USA 111, 3128–3133 (2014).
Oike, T. et al. A synthetic lethality–based strategy to treat cancers harboring a genetic deficiency in the chromatin remodeling factor BRG1. Cancer Res. 73, 5508–5518 (2013).
Ortmann, C.A. et al. Effect of mutation order on myeloproliferative neoplasms. N. Engl. J. Med. 372, 601–612 (2015).
Bultman, S. et al. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol. Cell 6, 1287–1295 (2000).
Hasselblatt, M. et al. SMARCA4-mutated atypical teratoid/rhabdoid tumors are associated with inherited germline alterations and poor prognosis. Acta Neuropathol. 128, 453–456 (2014).
Brennan, B., Stiller, C. & Bourdeaut, F. Extracranial rhabdoid tumours: what we have learned so far and future directions. Lancet Oncol. 14, e329–e336 (2013).
Schneppenheim, R. et al. Germline nonsense mutation and somatic inactivation of SMARCA4/BRG1 in a family with rhabdoid tumor predisposition syndrome. Am. J. Hum. Genet. 86, 279–284 (2010).
Foulkes, W.D. et al. No small surprise—small cell carcinoma of the ovary, hypercalcaemic type, is a malignant rhabdoid tumour. J. Pathol. 233, 209–214 (2014).
Govindan, R. et al. Genomic landscape of non–small cell lung cancer in smokers and never-smokers. Cell 150, 1121–1134 (2012).
Pomeroy, S.L. et al. Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature 415, 436–442 (2002).
Diskin, S.J. et al. STAC: a method for testing the significance of DNA copy number aberrations across multiple array-CGH experiments. Genome Res. 16, 1149–1158 (2006).
Trapnell, C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012).
Liu, Y., Hayes, D.N., Nobel, A. & Marron, J.S. Statistical significance of clustering for high-dimension, low–sample size data. J. Am. Stat. Assoc. 103, 1281–1293 (2008).
Rousseeuw, P.J. Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math. 20, 53–65 (1987).
Acknowledgements
We thank the following pathologists who provided archival material: C. Bazille, F. Beltjens, P.P. Bringuier, J. Calderaro, L. Chalabreysse, M.C. Charpentier, M.C. Château, A. Clemenson, S. Collardeau-Frachon, S. Croce, A. Croue, F. Dauchat, A.V. Decouvelaere, P. Dorfmuller, S. Duquenne, F. Forest, S. Garcia, M.R. Ghigna, I. Goubin Versini, A. Jouvet, E. Longchamp, P. Michenet, F. Mishellany, A. Moreau, H. Perrochia, B. PetitJean, J.M. Picquenot, P. Roger, V. Secques, I. Serre, I. Soubeyran, J.P. Terrier, A. Traverse-Glehen, I. Valo and L. Zemoura. We also thank the following clinicians who provided follow-up information: D. Arpin, F. Barlesi, J.O. Bay, T. Berghmans, P. Cassier, L. Cany, D. Cupissol, L. Diaz, P. Dumont, P. Fevrier, D. Fric, E. Huchot, N. Isambert, S. Labrune, V. Laurence, D. Moro-sibilot, M. Pays, I. Ray Coquard, O. Regnard, C. Rizzo, S. Salas and P.A. Thomas. We are grateful to J. Auclair, M. Carrere, S. Chabaud, A. Colombe, V. Dapremont, V. Haddad, M. Jean-Denis, F. Lorcy, N. Mesli, J.P. Michot, D. Postoly, G. Schummer, A. Shumikhin, V. Velascoa and Q. Wang. INSERM U830 is supported by the Institut National de la Santé et de la Recherche Médicale, the Institut Curie, the Ligue National Contre Le Cancer (Equipe Labellisée), the Institut National du Cancer and la Direction Générale de l'Offre de Soins (INCa-DGOS_5716), the European PROVABES (ERA-649 NET TRANSCAN JTC-2011), ASSET (FP7-HEALTH-2010-259348) and the Euro Ewing Consortium (HEALTH-F2-2013-602856) projects. W.R. is supported by the SiRIC Institut Curie program. U830 is also indebted to the Société Française des Cancers de l'Enfant, Enfants et Santé, Courir pour Mathieu, Dans les Pas du Géant, La Course de l'Espoir du Mont Valérien, Au Nom d'Andréa, Association Abigaël, Association Marabout de Ficelle, Les Bagouz à Manon, Les Amis de Claire and Association Adam. S.W. is supported by a grant from the Fondation Nuovo-Soldati. High-throughput sequencing was performed by the next-generation sequencing platform of Institut Curie, supported by grants ANR-10-EQPX-03 and ANR-10-INBS-09-08 from the Agence Nationale de la Recherche (Investissements d'Avenir) and by Canceropôle Ile-de-France. INSERM U1052 is supported by grants from LYRIC (DGOS-INCa 4664), NetSARC (INCa), RREPS (INCa), LabEx DEvweCAN (ANR-10-LABX-0061), Eurosarc (FP7-278742) and Ligue de l'Ain Contre le Cancer.
Author information
Authors and Affiliations
Contributions
F.L.L. initiated and designed the study, collected samples, performed experiments, analyzed the data and wrote the manuscript. S.W. initiated the study, analyzed the data and helped write the manuscript. G.P. and S. Ballet provided, prepared and sequenced biological samples of unclassified sarcoma. V.T.d.M., J.M.V., F.T.-B., D.R.-V., S.L. and F.G.-S. provided samples and were the reference senior pathologists. P.J.D., M.B., M.D.-S. and J.M.C. provided samples. N.F., E.F., B.B., G.Z. and N.G. recruited patients and provided clinical information. I.T. supervised all immunohistochemical experiments. D.P., S. Boyault and S.P. performed and analyzed OncoScan experiments. W.R., J.M.-P. and F.B. provided, prepared and sequenced biological samples of MRT and epithelioid sarcoma. Y.A. provided biological samples of RMC. A.A. and A.L. provided, prepared and sequenced biological samples of SCCOHT. O.D. gave biological and genetic guidance, provided infrastructure and helped write the manuscript. J.Y.B. initiated, designed and supervised the study and helped write the manuscript. F.T. initiated, designed, coordinated and supervised the study, performed the bioinformatics analyses, analyzed the data and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–24, Supplementary Tables 1, 3–11 and 14, and Supplementary Note. (PDF 8262 kb)
Supplementary Table 2
Variation identified by RNA-seq. (XLSX 113 kb)
Supplementary Table 12
Genes differentially expressed between SMARCA4-DTS and lung carcinoma (SMARCA4mt). (XLSX 127 kb)
Supplementary Table 13
Genes differentially expressed between SMARCA4-DTS and US(T). (XLSX 55 kb)
Rights and permissions
About this article
Cite this article
Le Loarer, F., Watson, S., Pierron, G. et al. SMARCA4 inactivation defines a group of undifferentiated thoracic malignancies transcriptionally related to BAF-deficient sarcomas. Nat Genet 47, 1200–1205 (2015). https://doi.org/10.1038/ng.3399
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3399
This article is cited by
-
The SMARCA4R1157W mutation facilitates chromatin remodeling and confers PRMT1/SMARCA4 inhibitors sensitivity in colorectal cancer
npj Precision Oncology (2023)
-
Thoracic SMARCA4-deficient undifferentiated tumor
Discover Oncology (2023)
-
Diagnosis and management of gastrointestinal SMARCA4-deficient undifferentiated tumors
Clinical Journal of Gastroenterology (2023)
-
Promising efficacy of immune checkpoint inhibitor plus chemotherapy for thoracic SMARCA4-deficient undifferentiated tumor
Journal of Cancer Research and Clinical Oncology (2023)
-
Molecular, clinicopathological characteristics and surgical results of resectable SMARCA4-deficient thoracic tumors
Journal of Cancer Research and Clinical Oncology (2023)