Abstract
No major predisposition gene for familial myeloproliferative neoplasms (MPN) has been identified. Here we demonstrate that the autosomal dominant transmission of a 700-kb duplication in four genetically related families predisposes to myeloid malignancies, including MPN, frequently progressing to leukemia. Using induced pluripotent stem cells and primary cells, we demonstrate that overexpression of ATG2B and GSKIP enhances hematopoietic progenitor differentiation, including of megakaryocytes, by increasing progenitor sensitivity to thrombopoietin (TPO). ATG2B and GSKIP cooperate with acquired JAK2, MPL and CALR mutations during MPN development. Thus, the germline duplication may change the fitness of cells harboring signaling pathway mutations and increases the probability of disease development.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Accession codes
References
Song, W.J. et al. Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develop acute myelogenous leukaemia. Nat. Genet. 23, 166–175 (1999).
Smith, M.L., Cavenagh, J.D., Lister, T.A. & Fitzgibbon, J. Mutation of CEBPA in familial acute myeloid leukemia. N. Engl. J. Med. 351, 2403–2407 (2004).
Hahn, C.N. et al. Heritable GATA2 mutations associated with familial myelodysplastic syndrome and acute myeloid leukemia. Nat. Genet. 43, 1012–1017 (2011).
Pasquet, M. et al. High frequency of GATA2 mutations in patients with mild chronic neutropenia evolving to MonoMac syndrome, myelodysplasia, and acute myeloid leukemia. Blood 121, 822–829 (2013).
Bluteau, D. et al. Thrombocytopenia-associated mutations in the ANKRD26 regulatory region induce MAPK hyperactivation. J. Clin. Invest. 124, 580–591 (2014).
Bellanné-Chantelot, C. et al. Genetic and clinical implications of the Val617Phe JAK2 mutation in 72 families with myeloproliferative disorders. Blood 108, 346–352 (2006).
Saint-Martin, C. et al. Analysis of the ten-eleven translocation 2 (TET2) gene in familial myeloproliferative neoplasms. Blood 114, 1628–1632 (2009).
Yamada, O. et al. Emergence of a BCR-ABL translocation in a patient with the JAK2V617F mutation: evidence for secondary acquisition of BCR-ABL in the JAK2V617F clone. J. Clin. Oncol. 32, e76–e79 (2014).
Jones, A.V. et al. JAK2 haplotype is a major risk factor for the development of myeloproliferative neoplasms. Nat. Genet. 41, 446–449 (2009).
Kilpivaara, O. et al. A germline JAK2 SNP is associated with predisposition to the development of JAK2V617F-positive myeloproliferative neoplasms. Nat. Genet. 41, 455–459 (2009).
Olcaydu, D. et al. A common JAK2 haplotype confers susceptibility to myeloproliferative neoplasms. Nat. Genet. 41, 450–454 (2009).
Jäger, R. et al. Common germline variation at the TERT locus contributes to familial clustering of myeloproliferative neoplasms. Am. J. Hematol. 89, 1107–1110 (2014).
Oddsson, A. et al. The germline sequence variant rs2736100_C in TERT associates with myeloproliferative neoplasms. Leukemia 28, 1371–1374 (2014).
Takahashi, K. et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 (2007).
Saliba, J. et al. Heterozygous and homozygous JAK2V617F states modeled by induced pluripotent stem cells from myeloproliferative neoplasm patients. PLoS ONE 8, e74257 (2013).
Takayama, N. et al. Generation of functional platelets from human embryonic stem cells in vitro via ES-sacs, VEGF-promoted structures that concentrate hematopoietic progenitors. Blood 111, 5298–5306 (2008).
Klimchenko, O. et al. A common bipotent progenitor generates the erythroid and megakaryocyte lineages in embryonic stem cell–derived primitive hematopoiesis. Blood 114, 1506–1517 (2009).
Vodyanik, M.A., Bork, J.A., Thomson, J.A. & Slukvin, I.I. Human embryonic stem cell–derived CD34+ cells: efficient production in the coculture with OP9 stromal cells and analysis of lymphohematopoietic potential. Blood 105, 617–626 (2005).
Prchal, J.F. & Axelrad, A.A. Letter: bone-marrow responses in polycythemia vera. N. Engl. J. Med. 290, 1382 (1974).
James, C. et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature 434, 1144–1148 (2005).
Jones, A.V. & Cross, N.C. Inherited predisposition to myeloproliferative neoplasms. Ther. Adv. Hematol. 4, 237–253 (2013).
Harutyunyan, A.S. & Kralovics, R. Role of germline genetic factors in MPN pathogenesis. Hematol. Oncol. Clin. North Am. 26, 1037–1051 (2012).
Krepischi, A.C., Pearson, P.L. & Rosenberg, C. Germline copy number variations and cancer predisposition. Future Oncol. 8, 441–450 (2012).
Kuiper, R.P., Ligtenberg, M.J., Hoogerbrugge, N. & Geurts van Kessel, A. Germline copy number variation and cancer risk. Curr. Opin. Genet. Dev. 20, 282–289 (2010).
Klampfl, T. et al. Genome integrity of myeloproliferative neoplasms in chronic phase and during disease progression. Blood 118, 167–176 (2011).
Rice, K.L. et al. Analysis of genomic aberrations and gene expression profiling identifies novel lesions and pathways in myeloproliferative neoplasms. Blood Cancer J. 1, e40 (2011).
Rumi, E. et al. Identification of genomic aberrations associated with disease transformation by means of high-resolution SNP array analysis in patients with myeloproliferative neoplasm. Am. J. Hematol. 86, 974–979 (2011).
Cui, W. et al. Trisomy 14 as a sole chromosome abnormality is associated with older age, a heterogenous group of myeloid neoplasms with dysplasia, and a wide spectrum of disease progression. J. Biomed. Biotechnol. 2010, 365318 (2010).
Mancini, M. et al. Trisomy 14 in hematologic diseases. Another non-random abnormality within myeloid proliferative disorders. Cancer Genet. Cytogenet. 66, 39–42 (1993).
Mertens, F. et al. Trisomy 14 in atypical chronic myeloid leukemia. Leukemia 4, 117–120 (1990).
Toze, C.L., Barnett, M.J., Naiman, S.C. & Horsman, D.E. Trisomy 14 is a non-random karyotypic abnormality associated with myeloid malignancies. Br. J. Haematol. 98, 177–185 (1997).
Bellanne-Chantelot, C., Jego, P., Lionne-Huyghe, P., Tulliez, M. & Najman, A. The JAK2V617F mutation may be present several years before the occurrence of overt myeloproliferative disorders. Leukemia 22, 450–451 (2008).
Rumi, E. et al. CALR exon 9 mutations are somatically acquired events in familial cases of essential thrombocythemia or primary myelofibrosis. Blood 123, 2416–2419 (2014).
Cabagnols, X., Cayuela, J.M. & Vainchenker, W. A CALR mutation preceding BCR-ABL1 in an atypical myeloproliferative neoplasm. N. Engl. J. Med. 372, 688–690 (2015).
Delhommeau, F. et al. Mutation in TET2 in myeloid cancers. N. Engl. J. Med. 360, 2289–2301 (2009).
Abdel-Wahab, O. et al. Genetic analysis of transforming events that convert chronic myeloproliferative neoplasms to leukemias. Cancer Res. 70, 447–452 (2010).
Lundberg, P. et al. Clonal evolution and clinical correlates of somatic mutations in myeloproliferative neoplasms. Blood 123, 2220–2228 (2014).
Yoshida, K. et al. The landscape of somatic mutations in Down syndrome–related myeloid disorders. Nat. Genet. 45, 1293–1299 (2013).
Chou, S.T. et al. Trisomy 21–associated defects in human primitive hematopoiesis revealed through induced pluripotent stem cells. Proc. Natl. Acad. Sci. USA 109, 17573–17578 (2012).
Maclean, G.A. et al. Altered hematopoiesis in trisomy 21 as revealed through in vitro differentiation of isogenic human pluripotent cells. Proc. Natl. Acad. Sci. USA 109, 17567–17572 (2012).
Gore, A. et al. Somatic coding mutations in human induced pluripotent stem cells. Nature 471, 63–67 (2011).
Genovese, G. et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl. J. Med. 371, 2477–2487 (2014).
Jaiswal, S. et al. Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 371, 2488–2498 (2014).
McKerrell, T. et al. Leukemia-associated somatic mutations drive distinct patterns of age-related clonal hemopoiesis. Cell Rep. 10, 1239–1245 (2015).
Xie, M. et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat. Med. 20, 1472–1478 (2014).
Lundberg, P. et al. Myeloproliferative neoplasms can be initiated from a single hematopoietic stem cell expressing JAK2-V617F. J. Exp. Med. 211, 2213–2230 (2014).
Kishi-Itakura, C., Koyama-Honda, I., Itakura, E. & Mizushima, N. Ultrastructural analysis of autophagosome organization using mammalian autophagy-deficient cells. J. Cell Sci. 127, 4089–4102 (2014).
Kang, M.R. et al. Frameshift mutations of autophagy-related genes ATG2B, ATG5, ATG9B and ATG12 in gastric and colorectal cancers with microsatellite instability. J. Pathol. 217, 702–706 (2009).
Mortensen, M., Watson, A.S. & Simon, A.K. Lack of autophagy in the hematopoietic system leads to loss of hematopoietic stem cell function and dysregulated myeloid proliferation. Autophagy 7, 1069–1070 (2011).
Warr, M.R. et al. FOXO3A directs a protective autophagy program in haematopoietic stem cells. Nature 494, 323–327 (2013).
Chou, H.Y. et al. GSKIP is homologous to the Axin GSK3 (interaction domain and functions as a negative regulator of GSK3β. Biochemistry 45, 11379–11389 (2006).
Lin, C.C. et al. GSKIP, an inhibitor of GSK3β, mediates the N-cadherin/β-catenin pool in the differentiation of SH-SY5Y cells. J. Cell. Biochem. 108, 1325–1336 (2009).
Li, D., August, S. & Woulfe, D.S. GSK3β is a negative regulator of platelet function and thrombosis. Blood 111, 3522–3530 (2008).
Soda, M., Willert, K., Kaushansky, K. & Geddis, A.E. Inhibition of GSK-3β promotes survival and proliferation of megakaryocytic cells through a β-catenin–independent pathway. Cell. Signal. 20, 2317–2323 (2008).
Abrahamsson, A.E. et al. Glycogen synthase kinase 3β missplicing contributes to leukemia stem cell generation. Proc. Natl. Acad. Sci. USA 106, 3925–3929 (2009).
Wang, Y. et al. The Wnt/β-catenin pathway is required for the development of leukemia stem cells in AML. Science 327, 1650–1653 (2010).
Tefferi, A. et al. Proposals and rationale for revision of the World Health Organization diagnostic criteria for polycythemia vera, essential thrombocythemia, and primary myelofibrosis: recommendations from an ad hoc international expert panel. Blood 110, 1092–1097 (2007).
Malak, S., Labopin, M., Saint-Martin, C., Bellanne-Chantelot, C. & Najman, A. Long term follow up of 93 families with myeloproliferative neoplasms: life expectancy and implications of JAK2V617F in the occurrence of complications. Blood Cells Mol. Dis. 49, 170–176 (2012).
Mali, P. et al. Improved efficiency and pace of generating induced pluripotent stem cells from human adult and fetal fibroblasts. Stem Cells 26, 1998–2005 (2008).
Debili, N. et al. Characterization of a bipotent erythro-megakaryocytic progenitor in human bone marrow. Blood 88, 1284–1296 (1996).
Livak, K.J. & Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 (2001).
Plo, I. et al. JAK2 stimulates homologous recombination and genetic instability: potential implication in the heterogeneity of myeloproliferative disorders. Blood 112, 1402–1412 (2008).
Acknowledgements
We greatly thank all the patients and family members involved in the study. We also thank O. Bawa and P. Opolon for the histopathological analysis of teratomas. We thank B. Benyahia for cytogenetic analysis. We thank M. Vestris for recruitment of patients. We greatly thank B. Job and the genomic platform for transcriptome and CGH analysis and also the iPSC platform of Institut Gustave Roussy. We thank S. Saker and T. Larmonier from the Généthon DNA and Cell Bank (Evry, France) for the establishment of B-lymphoblastoid cells from 'NMP' patients. We thank the cytometry platform of Institut Gustave Roussy (P. Rameau and Y. Lecluse). We thank C. Marzac for clinical data. We are grateful to S. Constantinescu and J. Feunteun for critical reading of the manuscript. We are also very grateful to E. Schwartz for proofreading the manuscript.
This work was supported by grants from Agence Nationale de la Recherche (ANR) (Blanc Megon 2009, Thrombocytosis 2011; ANR-13-JVSV1-GERMPN-01), Association pour la Recherche contre le Cancer (ARC) (Fondation ARC Libre 2012-SL220120605292), Groupe Information Santé (GIS)–Institute for Rare Diseases for High-Throughput Sequencing (AO9102LS), Association de Recherche sur la Moelle Osseuse (ARMO), regional Programme Hospitalier de Recherche Clinique (PHRC) AOR07014, Association Laurette Fugain and INCa-DGOS-INSERM 6043. Labex GR-Ex (I.P. and W.V.) is funded by the program 'Investissements d'Avenir'. G. Lenglet was supported by a postdoctoral fellowship from Ile-de-France Cancéropôle and ANR Molecular Medicine in Oncology (MMO) (funded by the program 'Investissements d'Avenir'). F.P. was supported by ARC. L.S. and J.S. were supported by doctoral grants from the Ile-de-France region (Cancéropôle and DIM Cellule Souche) and from Fondation pour la Recherche Médicale (FRM). C.M. was supported by ANR-13-JVSV1-GERMPN-01.
Author information
Authors and Affiliations
Contributions
W.V., I.P. and C.B.-C. designed and performed research, analyzed data, prepared figures and wrote the manuscript. J.S. performed research on iPSCs and primary cells. G. Lenglet, A.D.S. and L.S. performed research on iPSCs. C.S.-M., N. Droin and G. Leroy performed pangenomic analysis. C.M. performed immunoblot and qRT-PCR analyses. A.P. purified primary cells from donors. E.M. performed colony assays. F.P., J.-C.M., C.D.-D., P.F., F.I. and N.C. were involved in the clinical aspect of the study. A.N. was involved in the clinical aspect of the study and initiated the familial study of MPN. S.G. and S.C. were involved in the generation of EBVCs and their study. B.K. performed cytogenetic analysis. M'b.D., J.M. and P.D. carried out bioinformatics study of exome and transcriptomic data. N. Debili and H.R. provided experimental and/or intellectual input on iPSC culture and hematopoietic differentiation. V.D.V. performed TCL1 mouse modeling. E.S. and O.A.B. contributed intellectual input. All authors contributed to writing and editing.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
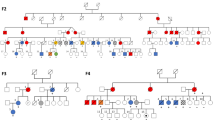
Supplementary Figure 1 A common CNV at 14q32 segregated with MPN in all families.
(a) A common haplotype at 14q32 segregated with MPN in both families. The haplotype of interest is depicted as a black bar. Sixteen microsatellite markers were genotyped: D14S81, D14S749, D14S1054, D14S1434, D14S1030, D14S265, D14S996, D14S987, D14S605, D14S131, D14S51, D14S979, D14S611, D14S1067, D14S65 and D14S267. Recombinations occurred in family F1 (patient F1:II.7) downstream of D14S265 (first marker excluded from the common haplotype) and in family F2 between D14S611 and D14S1067. (b) Gel electrophoresis showing the amplification of a 962-bp junction fragment in patients (F1:III-4, F2:III-10, F3:II-2 and F4:II-3) and its absence in non-carrier controls (F1:III-6, F2:IV-8 and F4:II-5). The primers used for PCR were Chr14_B2-C2.1F and Chr14_B1-C2.3R.
Supplementary Figure 2 Molecular characterization of P3-CNV-VF-TET2.
(a) Electropherograms of TET2 DNA sequences. (b) Measurements of the percentage of 5hmC levels by ELISA in P3-CNV-VF-TET2 versus P3-CNV-VF (one experiment). (c) Measurements of 5hmC/1000mC by liquid chromatography–mass spectrometry in erythroblasts from F2:II-4 compared to controls (results are the mean of three independent experiments ± s.e.m. for controls and one experiment for the patient).
Supplementary Figure 3 Molecular characterization of iPSCs.
(a) CGH array analysis is represented by hierarchical clustering with Pearson distance on 14 samples including iPSCs (a or b) and their respective CD34+ progenitor cells from control, P2, P3 and P4. Dots correspond to amplifications (in green, log2 (ratio) > 1.5) and deletions (in red, log2 (ratio) < –1.5), and lines correspond to gains (in blue, 0 < log2 (ratio) < 1.5) and losses (in red, 0 > log2 (ratio) > –1.5). Note that all iPSCs showed a CNV at the 14q locus, indicated by an arrow. As expected, the CGH array analysis showed that the 14q32.13-q32.2 duplication was present in P4-CNV, P3-CNV-VF and P3-CNV-VF-TET2 clones but did not identify significant acquired chromosome abnormalities in the iPSCs compared to the starting CD34+ progenitor cells. (b) Karyotypes of iPSCs from control, P4-CNV, P3-CNV-VF and P3-CNV-VF-TET2. iPSCs showed a normal karyotype except for a constitutional chromosomal abnormality inv(2)(p24.1q14.1) also observed in the primary cells of several patients from family F2 but absent from affected patients from families F1 and F4. Whole-exome sequencing analysis of iPSC clones compared to CD34+ starting cells identified a number of additional mutations compatible with previous reports, but none of these affected genes involved in hematological malignancies.
Supplementary Figure 4 Biological characterization of iPSCs.
(a) Analysis of alkaline phosphatase (AP) activity in iPSC colonies from P2-VF, P3-CNV-VF, P3-CNV-VF-TET2, P4-CNV or control. (b) Flow cytometry analysis of the TRA-1-81 and SSEA-4 pluripotency markers using specific antibodies or control immunoglobulin. (c) qRT-PCR performed for expression of the exogenous transgenes TgSOX2, TgOCT4, TgKLF4 and TgC-MYC in undifferentiated iPSCs using specific primers. PPIA was used as a housekeeping gene. Positive controls correspond to 293EBNA cells transduced with the retroviral vectors. (d) Fold change in exogenous transgenes (TgSOX2, TgOCT4, TgKLF4 and TgC-MYC) levels were compared to positive controls in iPSC-derived GPA+ cells by qRT-PCR (one experiment in duplicate). (e) Fold changes in endogenous pluripotent transcription factors (NANOG, POU5F1 and SOX2) in undifferentiated iPSCs were compared to ES cells by qRT-PCR. (f) The spontaneous differentiation of iPSCs and ES cells was induced by embryoid body formation. The presence of the three germ layers was assessed at day 5 by qRT-PCR performed on BRACHYURY (T), FOXA2 and PAX6. PPIA was used as a housekeeping gene. Results are expressed as fold change compared to the undifferentiated state (mean ± s.e.m., n = 4). (g) IPSCs were injected into immunodeficient NSG mice, and teratomas were analyzed after hematoxylin-eosin-saffron (HES) staining.
Supplementary Figure 5 Duplication modifies the sensitivity to EPO and TPO in iPSCs.
(a) GPA+CD41+ cells from P2-VF, P3-CNV-VF, P3-CNV-VF-TET2, P4-CNV or control were plated in methylcellulose in the presence of SCF and increasing concentrations of EPO. Erythroid progenitor colonies were counted 12 d later. (b) The percentage of large erythroid progenitor colonies (>50 cells per colony) was also calculated. Results are the mean ± s.e.m. of two experiments in duplicate. (c) Pictures of erythroid progenitors. (d) GPA+ cells were deprived of cytokines overnight in serum-free medium and seeded in IMDM alone for 4 h. Cells were stimulated or not with 10 U/ml EPO and analyzed by immunoblot. (e) CD41+ cells from P2-VF, P3-CNV-VF, P3-CNV-VF-TET2, P4-CNV or control were plated in plasma clots without or with increasing concentrations of TPO. CFU-MK colonies were counted at day 10 after indirect staining for CD41a. Results are the mean ± s.e.m. of two experiments in duplicate. (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). (f) CD41+ cells were sorted and grown, and the percentage of hyperploid cells (>8N) was calculated after propidium iodide labeling and flow cytometry analysis (mean ± s.e.m., n = 5; *P < 0.05, **P < 0.01). (g) Pictures of megakaryocyte cells in culture.
Supplementary Figure 6 Duplication is associated with autophagy.
(a) Platelets from healthy donors, patients or asymptomatic carriers were submitted to immunoblot analysis using LC3-I/II antibody. ß-actin was used as a loading control. Fold increases were calculated compared to control 1. (b) Relative fold change in WIPI1 expression in control EBV cell lines (n = 3 in triplicate) compared to patient cell lines (n = 5 in triplicate). (***P < 0.001.)
Supplementary Figure 7 Model of predisposition.
(a) In sporadic cases, essential thrombocythemia developed with a low frequency at a median age of 67 years as a result of the selection of a first hit (i.e., signaling mutation) either by aging or additional genetic or environmental events. Essential thrombocythemia (ET) can progress to myelofibrosis (MF). (b) In familial cases, the hematological malignancies or essential thrombocythemia developed with a high frequency at a median age of 41 years as a result of the CNV predisposition locus whose molecules cooperate with signaling mutations (JAK2 V617F) to change fitness and induce a growth advantage at the HSC/progenitor level. The progression is also more active and acute and is correlated with TET2 and/or IDH2 mutations.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 and Supplementary Tables 1–4. (PDF 1052 kb)
Rights and permissions
About this article
Cite this article
Saliba, J., Saint-Martin, C., Di Stefano, A. et al. Germline duplication of ATG2B and GSKIP predisposes to familial myeloid malignancies. Nat Genet 47, 1131–1140 (2015). https://doi.org/10.1038/ng.3380
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3380
This article is cited by
-
Driver mutation zygosity is a critical factor in predicting clonal hematopoiesis transformation risk
Blood Cancer Journal (2024)
-
MicroRNAs as the critical regulators of autophagy-mediated cisplatin response in tumor cells
Cancer Cell International (2023)
-
GSKIP modulates cell aggregation through EMT/MET signaling rather than differentiation in SH-SY5Y human neuroblastoma cells
Journal of Cell Communication and Signaling (2023)
-
Polymorphisms in autophagy genes are genetic susceptibility factors in glioblastoma development
BMC Cancer (2022)
-
The prognostic value of autophagy related genes with potential protective function in Ewing sarcoma
BMC Bioinformatics (2022)