Abstract
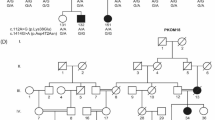
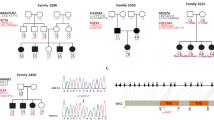
A duplication variant within the middle ear–specific gene A2ML1 cosegregates with otitis media in an indigenous Filipino pedigree (LOD score = 7.5 at reduced penetrance) and lies within a founder haplotype that is also shared by 3 otitis-prone European-American and Hispanic-American children but is absent in non-otitis-prone children and >62,000 next-generation sequences. We identified seven additional A2ML1 variants in six otitis-prone children. Collectively, our studies support a role for A2ML1 in the pathophysiology of otitis media.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Accession codes
References
Monasta, L. et al. Burden of disease caused by otitis media: systematic review and global estimates. PLoS ONE 7, e36226 (2012).
Acuin, J. Chronic Suppurative Otitis Media. Burden of Illness and Management Options (World Health Organization, 2004).
Casey, J.R. & Pichichero, M.E. Payment analysis of two diagnosis and management approaches of acute otitis media. Clin. Pediatr. (Phila.) 53, 865–873 (2014).
Kvestad, E. et al. Otitis media: genetic factors and sex differences. Twin Res. 7, 239–244 (2004).
Patel, J.A. et al. Association of proinflammatory cytokine gene polymorphisms with susceptibility to otitis media. Pediatrics 118, 2273–2279 (2006).
Daly, K.A. et al. Chronic and recurrent otitis media: a genome scan for susceptibility loci. Am. J. Hum. Genet. 75, 988–997 (2004).
Allen, E.K. et al. A genome-wide association study of chronic otitis media with effusion and recurrent otitis media identifies a novel susceptibility locus on chromosome 2. J. Assoc. Res. Otolaryngol. 14, 791–800 (2013).
Sakagami, M., Harada, T., Juhn, S.K. & Duvall, A.J. Morphologic and biochemical study of vascular permeability of the middle ear mucosa in experimental otitis media. Ann. Otol. Rhinol. Laryngol. 99, 654–659 (1990).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Marrero, A. et al. The crystal structure of human α2-macroglobulin reveals a unique molecular cage. Angew. Chem. Int. Edn Engl. 51, 3340–3344 (2012).
Wong, L.P. et al. Deep whole-genome sequencing of 100 southeast Asian Malays. Am. J. Hum. Genet. 92, 52–66 (2013).
Kircher, M. et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 46, 310–315 (2014).
Carlsson, B., Lundberg, C. & Ohlsson, K. Granulocyte protease inhibition in acute and chronic middle ear effusion. Acta Otolaryngol. (Stockh.) 95, 341–349 (1983).
Bondestam, M., Foucard, T. & Gebre-Medhin, M. Serum albumin, retinol-binding protein, thyroxin-binding prealbumin and acute phase reactants as indicators of undernutrition in children with undue susceptibility to acute infections. Acta Paediatr. Scand. 77, 94–98 (1988).
van Dongen, T.M., van der Heijden, G.J., Venekamp, R.P., Rovers, M.M. & Schilder, A.G. A trial of treatment for acute otorrhea in children with tympanostomy tubes. N. Engl. J. Med. 370, 723–733 (2014).
Marynen, P., Van Leuven, F., Cassiman, J.J. & Van den Berghe, H. Solubilization and affinity purification of the α2-macroglobulin receptor for human fibroblasts. J. Biol. Chem. 259, 7075–7079 (1984).
Vissers, L.E. et al. Heterozygous germline mutations in A2ML1 are associated with a disorder clinically related to Noonan syndrome. Eur. J. Hum. Genet. 23, 317–324 (2015).
HUGO Pan-Asian SNP Consortium. Mapping human genetic diversity in Asia. Science 326, 1541–1545 (2009).
Santos-Cortez, R.L.P. et al. Mutations in KARS, encoding lysyl-tRNA synthetase, cause autosomal-recessive nonsyndromic hearing impairment DFNB89. Am. J. Hum. Genet. 93, 132–140 (2013).
Wang, G.T., Peng, B. & Leal, S.M. Variant association tools for quality control and analysis of large-scale sequence and genotyping array data. Am. J. Hum. Genet. 94, 770–783 (2014).
Fishelson, M. & Geiger, D. Exact genetic linkage computations for general pedigrees. Bioinformatics 18 (suppl. 1), S189–S198 (2002).
Kelley, L.A. & Sternberg, M.J. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4, 363–371 (2009).
Reeve, J.P. & Rannala, B. DMLE+: Bayesian linkage disequilibrium gene mapping. Bioinformatics 18, 894–895 (2002).
Acknowledgements
We are very grateful to the indigenous community, families and individuals who participated in this study. We also thank M. Pedro, S.M. Lagrana, V. Ostan, and the surgical residents and audiology students who assisted in data collection and J. Belmont and S. Lalani for overall support. This study was supported by the Hearing Health Foundation, Action On Hearing Loss and the National Organization for Hearing Research Foundation (to R.L.P.S.-C.); the University of the Philippines Manila–National Institutes of Health (to G.T.A. and R.L.P.S.-C.); and US National Institutes of Health grants U54 HG006493 (to D.A.N.), R01 DK084350 (to M.M.S.), R01 DC003166 (to K.A.D.), R01 DC005841 (to T.C.), R01 DC011803 and R01 DC012564 (to S.R. and Z.M.A.), and R01 DC011651 and R01 DC003594 (to S.M.L.).
Author information
Authors and Affiliations
Consortia
Contributions
R.L.P.S.-C. and S.M.L. conceptualized the study. R.L.P.S.-C., C.M.C., M.R.T.R.-Q., M.L.C.T., M.C.G., E.G.D.V.L., P.J.L., T.L.I.G.-C., A.L.C., E.M.C.-d.l.P. and G.T.A. collected data and DNA samples from the indigenous population. J.A.P. and T.C. provided clinical data and DNA samples from the UTMB cohort. E.K.A., K.A.D. and M.M.S. provided clinical data and DNA samples from the UMN cohort. X.W., A.A., I.A. and R.L.P.S.-C. performed DNA isolation and PCR sequencing. The UWCMG, J.D.S., J.S., M.J.B. and D.A.N. performed genotyping and exome sequencing and provided exome data from non-otitis samples. J.D.S. performed principal-components analysis. G.T.W. and R.L.P.S.-C. performed exome analysis. B.L. estimated haplotype age. A.P.G., S.R. and Z.M.A. performed immunolocalization experiments. R.L.P.S.-C., S.R., Z.M.A. and S.M.L. wrote the manuscript. All authors read, provided critical input and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
A full list of members and affiliations appears in the Supplementary Note.
Integrated supplementary information
Supplementary Figure 1 Principal-components analysis for inferring ethnicity of 1,386 unrelated individuals whose DNA samples were submitted to UWCMG for exome sequencing.
Genotypes for common variants from UWCMG exomes were compared to HapMap 3 genotypes. ASW, African American; CEU, Utah/European American; CHB, Han Chinese; CHD, Chinese American; GIH, Gujarati Indian American; JPT, Japanese; LWK, Luhya Kenyan; MEX, Mexican American; MKK, Maasai Kenyan; TSI, Toscani Italian; YRI, Yoruba Nigerian. A large majority of 1,385 UWCMG individuals (blue crossed circles) who were used as controls for this study are of European or Hispanic American descent. One exome-sequenced individual from the indigenous Filipino population (red arrow) is positioned between the Mexican American and East Asian samples.
Supplementary Figure 2 Principal-components analysis (PCA) for inferring ethnicity of 41 unrelated UTMB children whose DNA samples were submitted for genome-wide genotyping.
Genotypes were compared to HapMap 3 genotypes (legend as in Supplementary Figure 1). Blue crossed circles denote UTMB children. The three otitis-prone children who carry the variant are indicated by their IDs. For these three children, self-reported ethnicity concurs with the PCA results. Of 38 non-otitis-prone children, 28 (73.7%) were confirmed to be European or Hispanic American, whereas 10 (26.3%) children were African American both by self-reporting and PCA. This ethnic distribution based on the availability of genome-wide genotypes is similar to that based on self-reported ethnicity for either the otitis-prone or non-otitis-prone group of UTMB children, i.e., 68% European or Hispanic American, 31% African American and 1% Asian American.
Supplementary Figure 3 Molecular modeling for the A2ML1 variants identified in the UTMB otitis-prone children.
(a) Stop-gain variants result in loss of thiol-ester and receptor-binding domains. (b) Variants p.Pro356Arg and p.Arg1001Trp are predicted to cause torsional changes and loss of hydrogen bonds. No obvious differences are due to p.Ala810Thr and p.Ala1431Val.
Supplementary Figure 4 A2ML1 has very faint or almost absent immunolocalization to the cochlea.
Whole-mount cochleae of P7 C57BL/6J mice immunolabeled with A2ML1 (green) showed very low or background levels of expression in the basal coil of the organ of Corti (OC). This very low or negative OC staining is consistent with no detectable mRNA expression for A2ML1 within the Shared Harvard Inner-Ear Laboratory Database. Actin (red) and nuclei/DAPI (blue) labeling were used to highlight the OC structure. Scale bar, 10 μm.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4, Supplementary Tables 1 and 2, and Supplementary Note. (PDF 1182 kb)
Rights and permissions
About this article
Cite this article
Santos-Cortez, R., Chiong, C., Reyes-Quintos, M. et al. Rare A2ML1 variants confer susceptibility to otitis media. Nat Genet 47, 917–920 (2015). https://doi.org/10.1038/ng.3347
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3347
This article is cited by
-
Cryo-EM structures of human A2ML1 elucidate the protease-inhibitory mechanism of the A2M family
Nature Communications (2022)
-
The clinical significance of A2ML1 variants in Noonan syndrome has to be reconsidered
European Journal of Human Genetics (2021)
-
Multi-omic studies on missense PLG variants in families with otitis media
Scientific Reports (2020)
-
Genetic counseling in an indigenous Filipino community with a high prevalence of A2ML1-related otitis media
Journal of Community Genetics (2019)
-
Recurrent Acute Otitis Media: What Are the Options for Treatment and Prevention?
Current Otorhinolaryngology Reports (2017)