Abstract
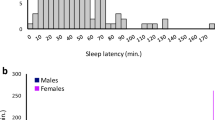
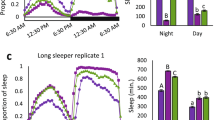
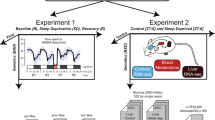
Sleep disorders are common in humans, and sleep loss increases the risk of obesity and diabetes1. Studies in Drosophila2,3 have revealed molecular pathways4,5,6,7 and neural tissues8,9,10 regulating sleep; however, genes that maintain genetic variation for sleep in natural populations are unknown. Here, we characterized sleep in 40 wild-derived Drosophila lines and observed abundant genetic variation in sleep architecture. We associated sleep with genome-wide variation in gene expression11 to identify candidate genes. We independently confirmed that molecular polymorphisms in Catsup (Catecholamines up) are associated with variation in sleep and that P-element mutations in four candidate genes affect sleep and gene expression. Transcripts associated with sleep grouped into biologically plausible genetically correlated transcriptional modules. We confirmed co-regulated gene expression using P-element mutants. Quantitative genetic analysis of natural phenotypic variation is an efficient method for revealing candidate genes and pathways.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Knutson, K.L. & Van Cauter, E. Associations between sleep loss and increased risk of obesity and diabetes. Ann. NY Acad. Sci. 1129, 287–304 (2008).
Hendricks, J.C. et al. Rest in Drosophila is a sleep-like state. Neuron 25, 129–138 (2000).
Shaw, P.J., Cirelli, C., Greenspan, R.J. & Tononi, G. Correlates of sleep and waking in Drosophila melanogaster. Science 287, 1834–1837 (2000).
Cirelli, C. et al. Reduced sleep in Drosophila Shaker mutants. Nature 434, 1087–1092 (2005).
Kume, K., Kume, S., Park, S.K., Hirsh, J. & Jackson, F.R. Dopamine is a regulator of arousal in the fruit fly. J. Neurosci. 25, 7377–7384 (2005).
Williams, J.A., Sathyanarayanan, S., Hendricks, J.C. & Sehgal, A. Interaction between sleep and the immune response in Drosophila: a role for the NFkB Relish. Sleep 30, 389–400 (2007).
Agosto, J. et al. Modulation of GABAA receptor desensitization uncouples sleep onset and maintenance in Drosophila. Nat. Neurosci. 11, 354–359 (2008).
Joiner, W.J., Crocker, A., White, B.H. & Sehgal, A. Sleep in Drosophila is regulated by adult mushroom bodies. Nature 441, 757–760 (2006).
Pitman, J.L., McGill, J.J., Keegan, K.P. & Allada, R. A dynamic role for the mushroom bodies in promoting sleep in Drosophila. Nature 441, 753–756 (2006).
Foltenyi, K., Greenspan, R.J. & Newport, J.W. Activation of EGFR and ERK by rhomboid signaling regulates the consolidation and maintenance of sleep in Drosophila. Nat. Neurosci. 10, 1160–1167 (2007).
Ayroles, J.F. et al. Systems genetics of complex traits in Drosophila melanogaster. Nat. Genet. advance online publication, doi:10.1038/ng.332 (22 February 2008).
Nitz, D.A., van Swinderen, B., Tononi, G. & Greenspan, R.J. Electrophysiological correlates of rest and activity in Drosophila melanogaster. Curr. Biol. 12, 1934–1940 (2002).
Tucker, A.M., Dinges, D.F. & Van Dongen, H.P.A. Trait interindividual differences in the sleep physiology of healthy young adults. J. Sleep Res. 16, 170–180 (2007).
Harbison, S.T. & Sehgal, A. Quantitative genetic analysis of sleep in Drosophila melanogaster. Genetics 178, 2341–2360 (2008).
Wu, M.N., Koh, K., Yue, Z., Joiner, W.J. & Sehgal, A. A genetic screen for sleep and circadian mutants reveals mechanisms underlying regulation of sleep in Drosophila. Sleep 31, 465–472 (2008).
Passador-Gurgel, G., Hsieh, W.-P., Hunt, P., Deighton, N. & Gibson, G. Quantitative trait transcripts for nicotine resistance in Drosophila melanogaster. Nat. Genet. 39, 264–268 (2007).
Cirelli, C., LaVaute, T.M. & Tononi, G. Sleep and wakefulness modulate gene expression in Drosophila. J. Neurochem. 94, 1411–1419 (2005).
Stathakis, D.G. et al. The Catecholamines up (Catsup) protein of Drosophila melanogaster functions as a negative regulator of tyrosine hydroxylase activity. Genetics 153, 361–382 (1999).
Andretic, R., van Swinderen, B. & Greenspan, R.J. Dopaminergic modulation of arousal in Drosophila. Curr. Biol. 15, 1165–1175 (2005).
Ganguly-Fitzgerald, I., Donlea, J. & Shaw, P.J. Waking experience affects sleep need in Drosophila. Science 313, 1775–1781 (2006).
O'Donnell, J.M., Stathakis, D.G., Burton, D.Y. & Chen, Z. Catecholamines-up, a negative regulator of tyrosine hydroxylase and GTP cyclohydrolase I in Drosophila melanogaster. in Chemistry and Biology of Pteridines and Folates (eds. Milstein, G. K. S., Levine, R. & Shane, B.) 211–215 (Kluwer Academic Publishers, Boston, 2002).
Bellen, H.J. et al. The BDGP Gene Disruption Project: single transposon insertions associated with 40% of Drosophila genes. Genetics 167, 761–781 (2004).
Dennis, G. et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4, R60 (2003).
Chintapalli, V.R., Wang, J. & Dow, J.A.T. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat. Genet. 39, 715–720 (2007).
Zhu, W. & Hanes, S.D. Identification of Drosophila Bicoid-interacting proteins using a custom two-hybrid selection. Gene 245, 329–339 (2000).
Sambandan, D., Yamamoto, A.H., Fanara, J.J., Mackay, T.F.C. & Anholt, R.R.H. Dynamic genetic interactions determine odor-guided behavior in Drosophila melanogaster. Genetics 174, 1349–1363 (2006).
Zepelin, H. Mammalian sleep. in Principles and Practice of Sleep Medicine (eds. Kryger, M. H., Roth, T. & Dement, W.C.) 69–80 (W.B. Saunders Company, Philadelphia, 1994).
Drosophila 12 Genomes Consortium. Evolution of genes and genomes on the Drosophila phylogeny. Nature 450, 203–218 (2007).
Larracuente, A.M. et al. Evolution of protein-coding genes in Drosophila. Trends Genet. 24, 114–123 (2008).
Acknowledgements
This work was supported by National Institutes of Health grants R01 GM 45146, R01 GM 076083 and R01 AA016560 to T.F.C.M. and the National Sleep Foundation Pickwick Fellowship to S.T.H. We thank D. Reif for assistance with Catsup permutation tests, K. Jordan for assistance with the genetic correlation data and R. Anholt for comments on the manuscript. This is a publication of the W.M. Keck Center for Behavioral Biology.
Author information
Authors and Affiliations
Contributions
S.T.H. and T.F.C.M. conceived of the experiment. S.T.H measured sleep phenotypes in all lines and measured expression levels in P-element insertions. M.A.C. generated whole-genome expression data and sequenced Catsup. S.T.H, J.F.A. and E.A.S. analyzed the data. R.F.L. generated the 40-line reference panel. S.T.H. and T.F.C.M wrote the paper.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1,3,4 and 6, Supplementary Figure 1 and Supplementary Methods (PDF 940 kb)
Supplementary Table 2
SFPs and modules of correlated transcripts associated with quantitative sleep traits (XLS 541 kb)
Supplementary Table 5
GO categories (XLS 151 kb)
Rights and permissions
About this article
Cite this article
Harbison, S., Carbone, M., Ayroles, J. et al. Co-regulated transcriptional networks contribute to natural genetic variation in Drosophila sleep. Nat Genet 41, 371–375 (2009). https://doi.org/10.1038/ng.330
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.330
This article is cited by
-
Rethinking Neuroscientific Methodology: Lived Experience in Behavioral Studies
Biological Theory (2024)
-
Zinc antagonizes iron-regulation of tyrosine hydroxylase activity and dopamine production in Drosophila melanogaster
BMC Biology (2021)
-
Phenotypic coupling of sleep and starvation resistance evolves in D. melanogaster
BMC Evolutionary Biology (2020)
-
Evidence that natural selection maintains genetic variation for sleep in Drosophila melanogaster
BMC Evolutionary Biology (2015)
-
Characterizing the neurotranscriptomic states in alternative stress coping styles
BMC Genomics (2015)