Abstract
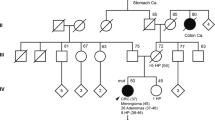
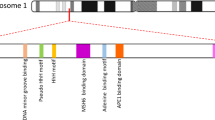
The genetic cause underlying the development of multiple colonic adenomas, the premalignant precursors of colorectal cancer (CRC), frequently remains unresolved in patients with adenomatous polyposis. Here we applied whole-exome sequencing to 51 individuals with multiple colonic adenomas from 48 families. In seven affected individuals from three unrelated families, we identified a homozygous germline nonsense mutation in the base-excision repair (BER) gene NTHL1. This mutation was exclusively found in a heterozygous state in controls (minor allele frequency of 0.0036; n = 2,329). All three families showed recessive inheritance of the adenomatous polyposis phenotype and progression to CRC in at least one member. All three affected women developed an endometrial malignancy or premalignancy. Genetic analysis of three carcinomas and five adenomas from different affected individuals showed a non-hypermutated profile enriched for cytosine-to-thymine transitions. We conclude that a homozygous loss-of-function germline mutation in the NTHL1 gene predisposes to a new subtype of BER-associated adenomatous polyposis and CRC.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Greenman, C. et al. Patterns of somatic mutation in human cancer genomes. Nature 446, 153–158 (2007).
Al-Tassan, N. et al. Inherited variants of MYH associated with somatic G:C→T:A mutations in colorectal tumors. Nat. Genet. 30, 227–232 (2002).
Palles, C. et al. Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat. Genet. 45, 136–144 (2013).
Briggs, S. & Tomlinson, I. Germline and somatic polymerase ɛ and δ mutations define a new class of hypermutated colorectal and endometrial cancers. J. Pathol. 230, 148–153 (2013).
Lipton, L. et al. Carcinogenesis in MYH-associated polyposis follows a distinct genetic pathway. Cancer Res. 63, 7595–7599 (2003).
Mongin, C. et al. Unexplained polyposis: a challenge for geneticists, pathologists and gastroenterologists. Clin. Genet. 81, 38–46 (2012).
Rahman, N. Realizing the promise of cancer predisposition genes. Nature 505, 302–308 (2014).
Dallosso, A.R. et al. Inherited predisposition to colorectal adenomas caused by multiple rare alleles of MUTYH but not OGG1, NUDT1, NTH1 or NEIL 1, 2 or 3. Gut 57, 1252–1255 (2008).
Smith, C.G. et al. Role of the oxidative DNA damage repair gene OGG1 in colorectal tumorigenesis. J. Natl. Cancer Inst. 105, 1249–1253 (2013).
Broderick, P. et al. Evaluation of NTHL1, NEIL1, NEIL2, MPG, TDG, UNG and SMUG1 genes in familial colorectal cancer predisposition. BMC Cancer 6, 243 (2006).
Nielsen, M. et al. Colorectal carcinomas in MUTYH-associated polyposis display histopathological similarities to microsatellite unstable carcinomas. BMC Cancer 9, 184 (2009).
Cooke, M.S., Evans, M.D., Dizdaroglu, M. & Lunec, J. Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J. 17, 1195–1214 (2003).
Chan, M.K. et al. Targeted deletion of the genes encoding NTH1 and NEIL1 DNA N-glycosylases reveals the existence of novel carcinogenic oxidative damage to DNA. DNA Repair (Amst.) 8, 786–794 (2009).
Cancer Genome Atlas Research Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337 (2012).
Corral, R. et al. Genetic variation in the base excision repair pathway, environmental risk factors, and colorectal adenoma risk. PLoS ONE 8, e71211 (2013).
Gilissen, C. et al. Exome sequencing identifies WDR35 variants involved in Sensenbrenner syndrome. Am. J. Hum. Genet. 87, 418–423 (2010).
International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature 449, 851–861 (2007).
Carr, I.M. et al. Autozygosity mapping with exome sequence data. Hum. Mutat. 34, 50–56 (2013).
Kandoth, C. et al. Mutational landscape and significance across 12 major cancer types. Nature 502, 333–339 (2013).
O'Roak, B.J. et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science 338, 1619–1622 (2012).
Boyle, E.A., O'Roak, B.J., Martin, B.K., Kumar, A. & Shendure, J. MIPgen: optimized modeling and design of molecular inversion probes for targeted resequencing. Bioinformatics 30, 2670–2672 (2014).
Hiatt, J.B., Pritchard, C.C., Salipante, S.J., O'Roak, B.J. & Shendure, J. Single molecule molecular inversion probes for targeted, high-accuracy detection of low-frequency variation. Genome Res. 23, 843–854 (2013).
Acknowledgements
We thank N. Arts, A. van Raaij, S. van Vliet, M. van de Vorst and M. Kwint for technical assistance, and R. Derks and M. Nelen for targeted tumor sequencing and data analysis. We thank H. Brunner for critical reading of the manuscript. The authors would like to thank the Exome Aggregation Consortium and the groups that provided exome variant data for comparison. A full list of contributing groups can be found at http://exac.broadinstitute.org/about. This work was supported by research grants from the Dutch Cancer Society (KWF; grant 2009-4335) and the Netherlands Organization for Scientific Research (NWO; grant 917-10-358).
Author information
Authors and Affiliations
Contributions
R.D.A.W., R.P.K. and N.H. designed the study. R.D.A.W. analyzed and interpreted whole-exome sequencing data. R.D.A.W., R.M.d.V. and E.J.K. performed laboratory experiments and/or analyzed data. E.T.P.V. and J.Y.H.-K. performed haploblock analysis. R.D.A.W. and B.B.J.T. performed somatic mutation analyses. C.G. supervised bioinformatics data assembly and performed statistical analysis. J.S., E.A.B. and A.H. designed molecular inversion probes. I.D.N. and J.H.J.M.v.K. collected tumor samples and interpreted histology. C.M.K., L.S., W.A.G.v.Z.-S., M.C.J., F.M.N. and N.H. were responsible for patient counseling and clinical data acquisition. M.J.L.L., A.G.v.K., R.P.K. and N.H. supervised the work. R.D.A.W., R.P.K. and N.H. wrote the manuscript, with assistance and final approval from all co-authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–10 and Supplementary Figures 1–6. (PDF 2347 kb)
Rights and permissions
About this article
Cite this article
Weren, R., Ligtenberg, M., Kets, C. et al. A germline homozygous mutation in the base-excision repair gene NTHL1 causes adenomatous polyposis and colorectal cancer. Nat Genet 47, 668–671 (2015). https://doi.org/10.1038/ng.3287
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3287
This article is cited by
-
Curative resection via right hemicolectomy and regional lymph node dissection for colonic adenomatous polyposis of unknown etiology with adenocarcinomas localized in the right side of the colon: a case report
Surgical Case Reports (2024)
-
Founder pathogenic variants in colorectal neoplasia susceptibility genes in Ashkenazi Jews undergoing colonoscopy
BJC Reports (2024)
-
Sequence variants affecting the genome-wide rate of germline microsatellite mutations
Nature Communications (2023)
-
Genotype–Phenotype Correlations in Autosomal Dominant and Recessive APC Mutation-Negative Colorectal Adenomatous Polyposis
Digestive Diseases and Sciences (2023)
-
A large family with MSH3-related polyposis
Familial Cancer (2023)