Abstract
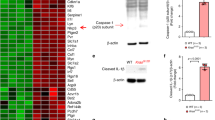
Glucocorticoids are universally used in the treatment of acute lymphoblastic leukemia (ALL), and resistance to glucocorticoids in leukemia cells confers poor prognosis. To elucidate mechanisms of glucocorticoid resistance, we determined the prednisolone sensitivity of primary leukemia cells from 444 patients newly diagnosed with ALL and found significantly higher expression of CASP1 (encoding caspase 1) and its activator NLRP3 in glucocorticoid-resistant leukemia cells, resulting from significantly lower somatic methylation of the CASP1 and NLRP3 promoters. Overexpression of CASP1 resulted in cleavage of the glucocorticoid receptor, diminished the glucocorticoid-induced transcriptional response and increased glucocorticoid resistance. Knockdown or inhibition of CASP1 significantly increased glucocorticoid receptor levels and mitigated glucocorticoid resistance in CASP1-overexpressing ALL. Our findings establish a new mechanism by which the NLRP3-CASP1 inflammasome modulates cellular levels of the glucocorticoid receptor and diminishes cell sensitivity to glucocorticoids. The broad impact on the glucocorticoid transcriptional response suggests that this mechanism could also modify glucocorticoid effects in other diseases.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Yudt, M.R. & Cidlowski, J.A. The glucocorticoid receptor: coding a diversity of proteins and responses through a single gene. Mol. Endocrinol. 16, 1719–1726 (2002).
Pui, C.H. et al. Treating childhood acute lymphoblastic leukemia without cranial irradiation. N. Engl. J. Med. 360, 2730–2741 (2009).
Den Boer, M.L. et al. Patient stratification based on prednisolone-vincristine-asparaginase resistance profiles in children with acute lymphoblastic leukemia. J. Clin. Oncol. 21, 3262–3268 (2003).
Kaspers, G.J. et al. In vitro cellular drug resistance and prognosis in newly diagnosed childhood acute lymphoblastic leukemia. Blood 90, 2723–2729 (1997).
Pieters, R. et al. Relation of cellular drug resistance to long-term clinical outcome in childhood acute lymphoblastic leukaemia. Lancet 338, 399–403 (1991).
Dördelmann, M. et al. Prednisone response is the strongest predictor of treatment outcome in infant acute lymphoblastic leukemia. Blood 94, 1209–1217 (1999).
Wellington, C.L. et al. Caspase cleavage of gene products associated with triplet expansion disorders generates truncated fragments containing the polyglutamine tract. J. Biol. Chem. 273, 9158–9167 (1998).
Boxer, M.B., Shen, M., Auld, D.S., Wells, J.A. & Thomas, C.J. A small molecule inhibitor of Caspase 1. Probe Reports from the NIH Molecular Libraries Program, http://www.ncbi.nlm.nih.gov/books/NBK56241/ (2010).
Ogura, Y., Sutterwala, F.S. & Flavell, R.A. The inflammasome: first line of the immune response to cell stress. Cell 126, 659–662 (2006).
Mariathasan, S. et al. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature 440, 228–232 (2006).
Shenoy, A.R. et al. GBP5 promotes NLRP3 inflammasome assembly and immunity in mammals. Science 336, 481–485 (2012).
Lu, B. et al. Novel role of PKR in inflammasome activation and HMGB1 release. Nature 488, 670–674 (2012).
Subramanian, N., Natarajan, K., Clatworthy, M.R., Wang, Z. & Germain, R.N. The adaptor MAVS promotes NLRP3 mitochondrial localization and inflammasome activation. Cell 153, 348–361 (2013).
Reddy, T.E. et al. Genomic determination of the glucocorticoid response reveals unexpected mechanisms of gene regulation. Genome Res. 19, 2163–2171 (2009).
Meijsing, S.H. et al. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science 324, 407–410 (2009).
ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Hogan, L.E. et al. Integrated genomic analysis of relapsed childhood acute lymphoblastic leukemia reveals therapeutic strategies. Blood 118, 5218–5226 (2011).
Ray, C.A. et al. Viral inhibition of inflammation: cowpox virus encodes an inhibitor of the interleukin-1β converting enzyme. Cell 69, 597–604 (1992).
Komiyama, T. et al. Inhibition of interleukin-1β converting enzyme by the cowpox virus serpin CrmA. An example of cross-class inhibition. J. Biol. Chem. 269, 19331–19337 (1994).
Garcia-Calvo, M. et al. Inhibition of human caspases by peptide-based and macromolecular inhibitors. J. Biol. Chem. 273, 32608–32613 (1998).
Wang, J.C. et al. Chromatin immunoprecipitation (ChIP) scanning identifies primary glucocorticoid receptor target genes. Proc. Natl. Acad. Sci. USA 101, 15603–15608 (2004).
Charmandari, E. et al. A novel point mutation in helix 11 of the ligand-binding domain of the human glucocorticoid receptor gene causing generalized glucocorticoid resistance. J. Clin. Endocrinol. Metab. 92, 3986–3990 (2007).
Pui, C.H., Dahl, G.V., Rivera, G., Murphy, S.B. & Costlow, M.E. The relationship of blast cell glucocorticoid receptor levels to response to single-agent steroid trial and remission response in children with acute lymphoblastic leukemia. Leuk. Res. 8, 579–585 (1984).
Houghton, P.J. et al. The pediatric preclinical testing program: description of models and early testing results. Pediatr. Blood Cancer 49, 928–940 (2007).
Shoemaker, R.H. The NCI60 human tumour cell line anticancer drug screen. Nat. Rev. Cancer 6, 813–823 (2006).
Bachmann, P.S., Gorman, R., Mackenzie, K.L., Lutze-Mann, L. & Lock, R.B. Dexamethasone resistance in B-cell precursor childhood acute lymphoblastic leukemia occurs downstream of ligand-induced nuclear translocation of the glucocorticoid receptor. Blood 105, 2519–2526 (2005).
Bachmann, P.S. et al. Divergent mechanisms of glucocorticoid resistance in experimental models of pediatric acute lymphoblastic leukemia. Cancer Res. 67, 4482–4490 (2007).
Holleman, A. et al. Gene-expression patterns in drug-resistant acute lymphoblastic leukemia cells and response to treatment. N. Engl. J. Med. 351, 533–542 (2004).
Pottier, N. et al. The SWI/SNF chromatin-remodeling complex and glucocorticoid resistance in acute lymphoblastic leukemia. J. Natl. Cancer Inst. 100, 1792–1803 (2008).
Jones, C.L. et al. Loss of TBL1XR1 disrupts glucocorticoid receptor recruitment to chromatin and results in glucocorticoid resistance in a B-lymphoblastic leukemia model. J. Biol. Chem. 289, 20502–20515 (2014).
Bialer, M. et al. Progress report on new antiepileptic drugs: a summary of the Eleventh Eilat Conference (EILAT XI). Epilepsy Res. 103, 2–30 (2013).
Schroder, K. & Tschopp, J. The inflammasomes. Cell 140, 821–832 (2010).
Zhou, R., Tardivel, A., Thorens, B., Choi, I. & Tschopp, J. Thioredoxin-interacting protein links oxidative stress to inflammasome activation. Nat. Immunol. 11, 136–140 (2010).
Martinon, F., Petrilli, V., Mayor, A., Tardivel, A. & Tschopp, J. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature 440, 237–241 (2006).
Karl, M. et al. Familial glucocorticoid resistance caused by a splice site deletion in the human glucocorticoid receptor gene. J. Clin. Endocrinol. Metab. 76, 683–689 (1993).
Bouligand, J. et al. Familial glucocorticoid receptor haploinsufficiency by non-sense mediated mRNA decay, adrenal hyperplasia and apparent mineralocorticoid excess. PLoS ONE 5, e13563 (2010).
Michailidou, Z. et al. Glucocorticoid receptor haploinsufficiency causes hypertension and attenuates hypothalamic-pituitary-adrenal axis and blood pressure adaptions to high-fat diet. FASEB J. 22, 3896–3907 (2008).
Riml, S., Schmidt, S., Ausserlechner, M.J., Geley, S. & Kofler, R. Glucocorticoid receptor heterozygosity combined with lack of receptor auto-induction causes glucocorticoid resistance in Jurkat acute lymphoblastic leukemia cells. Cell Death Differ. 11 (suppl 1) S65–S72 (2004).
McKay, L.I. & Cidlowski, J.A. Cross-talk between nuclear factor–κB and the steroid hormone receptors: mechanisms of mutual antagonism. Mol. Endocrinol. 12, 45–56 (1998).
Teurich, S. & Angel, P. The glucocorticoid receptor synergizes with Jun homodimers to activate AP-1–regulated promoters lacking GR binding sites. Chem. Senses 20, 251–255 (1995).
Hubbell, E., Liu, W.M. & Mei, R. Robust estimators for expression analysis. Bioinformatics 18, 1585–1592 (2002).
Gautier, L., Cope, L., Bolstad, B.M. & Irizarry, R.A. affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20, 307–315 (2004).
Gentleman, R.C. et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5, R80 (2004).
Cheok, M.H. et al. Treatment-specific changes in gene expression discriminate in vivo drug response in human leukemia cells. Nat. Genet. 34, 85–90 (2003).
Yeoh, E.J. et al. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell 1, 133–143 (2002).
Kuan, P.F., Wang, S., Zhou, X. & Chu, H. A statistical framework for Illumina DNA methylation arrays. Bioinformatics 26, 2849–2855 (2010).
Stouffer, S.A., Suchman, E.A., DeVinney, L.C., Star, S.A. & Williams, R.M. Jr. The American Soldier: Adjustment during Army Life (Studies in Social Psychology in World War II, Vol. 1.) (Princeton University Press, 1949).
Yang, X. et al. A public genome-scale lentiviral expression library of human ORFs. Nat. Methods 8, 659–661 (2011).
Boehm, J.S. et al. Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell 129, 1065–1079 (2007).
Muzio, M., Salvesen, G.S. & Dixit, V.M. FLICE induced apoptosis in a cell-free system. Cleavage of caspase zymogens. J. Biol. Chem. 272, 2952–2956 (1997).
Savic, D., Gertz, J., Jain, P., Cooper, G.M. & Myers, R.M. Mapping genome-wide transcription factor binding sites in frozen tissues. Epigenetics Chromatin 6, 30 (2013).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Bailey, T.L., Williams, N., Misleh, C. & Li, W.W. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34, W369–W373 (2006).
Acknowledgements
We gratefully acknowledge the patients and their parents who participated in this research. We are appreciative of the expert technical assistance of M. Roberts, Y. Chu, Y. Wang, M.A. Payton, J. Stukenborg, S. Salehy, M. Needham, M. Chung, N. Lenchik, M. Loyd and E. Walker. We thank J. Groff and E. Stevens for figure preparation assistance and C. Simmons for assistance with manuscript preparation. We thank D. Green for his scientific advice and discussion of the manuscript. We thank C. Stewart, G. Neale, J. Morris and K. Rakestraw for their technical advice and expertise. This work was supported in part by US National Institutes of Health (NIH) National Cancer Institute grant R37CA36401 (W.E.E., M.V.R. and C.-H.P.), US NIH National Institute of General Medical Sciences Pharmacogenomics Research Network grant U01GM92666 (M.V.R. and W.E.E.), US NIH grant F32CA141762 (S.W.P.) and an American Recovery and Reinvestment Act supplement, 3R37CA036401-26S1 (W.E.E.). This work was also supported by Cancer Center Support Grant CA21765 from the National Cancer Institute and by the American Lebanese Syrian Associated Charities (ALSAC). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
S.W.P., E.J.B., D.S., L.B.R., W.E.T., P.G., R.K.S.M., M.A., A.M., J.M., D.R.C., L.T.L., Y.F., R.K.G., T.-D.K., M.V.R. and W.E.E. designed experiments. C.-H.P., S.J., M.V.R. and W.E.E. designed clinical trials. S.W.P., E.J.B., D.S., L.B.R., W.E.T., P.G., R.K.S.M., M.A., A.M., D.R.C., L.T.L., Y.F., A.Z., A.G., D.C., J.J.B. and L.H. performed experiments. S.W.P., E.J.B., D.S., L.B.R., W.E.T. and W.E.E. wrote the manuscript (reviewed by all authors). S.W.P., E.J.B., D.S., L.B.R., W.E.T., D.R.C., L.T.L., J.C.P., J.R.M., Y.F., K.R.C., G.S., M.R.W., A.M.F., C.C., W.Y., S.E.K., C.A.F., B.D., C.S., J.K.H., A.Z., A.G., D.C., J.J.B., L.H., C.G.M., M.L.d.B., R.P., S.J., T.L.D., F.L., D.B., W.L.C., C.-H.P., R.M.M., R.K.G., T.-D.K., M.V.R. and W.E.E. analyzed data.
Corresponding author
Ethics declarations
Competing interests
W.E.E., S.W.P. and E.J.B. are named as co-inventors on a pending patent application that relates to the subject matter of the article, which was filed by St. Jude Children's Research Hospital.
Integrated supplementary information
Supplementary Figure 1 Glucocorticoid-resistant leukemia cells have higher expression of CASP1 and NLRP3.
CASP1 (a–c) and NLRP3 (d–f) expression was significantly higher in glucocorticoid-resistant leukemia cells from three cohorts of newly diagnosed patients. Exact Wilcoxon Mann-Whitney rank-sum test P values are shown for a–f, with Stouffer's z-score method meta-analysis P values shown above a–f as described in the Online Methods.
Supplementary Figure 2 CASP1 and NLRP3 methylation probe locations.
DNA methylation analysis probe locations for CASP1 and NLRP3 relative to the transcription start sites of these genes. The specific base analyzed is shown in square brackets with the genomic context surrounding each site.
Supplementary Figure 3 Hypomethylation of the CASP1 and NLRP3 promoter region was associated with higher CASP1 and NLRP3 expression in leukemia cells.
In both patient cohorts for which DNA was available for DNA methylation analysis (St. Jude Protocols XV and XVI), significantly lower levels of CASP1 (a,b) and NLRP3 (d,e) methylation were found in leukemia cells with higher expression of CASP1 and NLRP3. For both CASP1 and NLRP3 methylation status, the DNA methylation site (CpG) was within 100 bp of the transcription start site (Supplementary Fig. 2). k-means clustering analysis (a triangle represents k means–identified group A, a circle represents k means–identified group B, pink and red squares represent k means–identified centers for group A and B, respectively) utilizing only CASP1 and NLRP3 methylation status significantly discriminated sensitive leukemias (blue symbols; higher methylation) from resistant leukemias (orange symbols; lower methylation) in patients from both St. Jude Protocol XV and St. Jude Protocol XVI (g,h). Exact Wilcoxon Mann-Whitney rank-sum test P values are shown for a, b, d and e, with Stouffer's z-score method meta-analysis P values shown above a, b, d and e.
Supplementary Figure 4 Germline versus somatic DNA methylation status.
In a subset of patients (n = 55) enrolled on St. Jude Protocol XVI, both germline and somatic DNA methylation was analyzed. (a,b) DNA methylation in these patients, with lines connecting paired samples. Orange symbols show prednisolone-resistant patients, blue symbols show prednisolone-sensitive patients and gray symbols show patients with intermediate resistance as defined in Fig. 1c. Paired t-test P values are shown for a and b.
Supplementary Figure 5 CASP1 increases resistance to glucocorticoids.
Enforced expression of CASP1 in a human B-lineage leukemia cell line (697; harboring an E2A-PBX1 translocation) increased resistance to prednisolone after activation of the NALP3 inflammasome (by addition of LPS and ATP). 697 cells were transduced with a lentivirus containing full-length CASP1 and genes encoding puromycin N-acetyltransferase or puromycin N-acetyltransferase alone (control). Cells were selected with puromycin, and their sensitivity to prednisolone was measured using the MTT assay, in the presence (+) or absence (–) of inflammasome activation (LPS and ATP). LC50 values for control and CASP1-expressing cells in the presence of LPS and ATP were 0.73 ± 0.16 µM and 1.9 ± 0.73 µM prednisolone, respectively (P = 0.01).
Supplementary Figure 6 CrmA expression increases glucocorticoid receptor levels and restores glucocorticoid sensitivity in primary glucocorticoid-resistant leukemia cells.
Primary leukemia cells isolated from a patient with glucocorticoid-resistant ALL and high levels of CASP1 expression were transduced with lentivirus either expressing RFP plus the CASP1-inhibitory protein CrmA (CrmA) or RFP alone. (a) The sensitivity (LC50) of these cells to dexamethasone was determined by MTT assay, revealing that overexpression of CrmA reversed glucocorticoid resistance (LC50 = 0.14 μM (95% confidence interval = 0.1426 × 10–2 to 0.2773 μM versus >10 μM; P < 1 × 10–14)). (b) Western blot analysis demonstrates restoration of glucocorticoid receptor protein expression.
Supplementary Figure 7 Endogenous CASP1 protein levels in glucocorticoid-resistant primary ALLs are comparable to CASP1 protein level in the NALM-6 leukemia cells expressing recombinant CASP1.
CASP1 protein levels by western blot in primary leukemia cells from two glucocorticoid-resistant patients and NALM-6 cells expressing recombinant CASP1 are shown. Lane 1 is lysate from NALM-6 ALL cells transduced with the control (empty) lentivirus and has undetectable CASP1, lane 2 depicts lysate from CASP1-transduced NALM-6 leukemia cells (in which CASP1 was not activated with LPS and ATP), and lanes 3 and 4 are lysates from primary leukemia cells isolated from patients with glucocorticoid-resistant ALL (LC50 = 1,387 and 206.4 μM, respectively). Recombinant CASP1 has a higher molecular weight than endogenous CASP1, owing to the Myc-DDK protein tag.
Supplementary Figure 8 Glucocorticoid-resistant cell lines derived from ALL patient xenografts and NCI60 cell lines often show increased expression of CASP1.
Publically available data from xenografts generated from glucocorticoid-resistant and -sensitive primary leukemia samples (Online Methods) were analyzed for CASP1 expression. (a) Xenografts that were resistant to dexamethasone showed higher CASP1 expression than those that were responsive to dexamethasone, including mRNA expression data from two glucocorticoid-resistant ALL xenografts established from newly diagnosed patients at St. Jude Children’s Research Hospital. (b) In a separate analysis, the REH and Nalm6 cell lines and ALL cells in the NCI60 leukemia panel were combined to compare CASP1 expression in glucocorticoid-sensitive and -resistant ALLs. Similar to the xenograft data (a), cell lines that were resistant to prednisolone had higher expression of CASP1. Combining these data revealed a highly significant difference in CASP1 expression levels in glucocorticoid-sensitive and glucocoritcoid-resistant cell lines and xenografts (Wilcoxon rank-sum test, P = 8.9 × 10–7).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8 and Supplementary Tables 1 and 5. (PDF 466 kb)
Supplementary Table 2
Upregulated genes. (XLS 60 kb)
Supplementary Table 3
Downregulated genes. (XLS 67 kb)
Supplementary Table 4
Random unchanged genes. (XLS 488 kb)
Rights and permissions
About this article
Cite this article
Paugh, S., Bonten, E., Savic, D. et al. NALP3 inflammasome upregulation and CASP1 cleavage of the glucocorticoid receptor cause glucocorticoid resistance in leukemia cells. Nat Genet 47, 607–614 (2015). https://doi.org/10.1038/ng.3283
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3283
This article is cited by
-
The role of inflammasomes in human diseases and their potential as therapeutic targets
Signal Transduction and Targeted Therapy (2024)
-
Immune mechanisms of depression in rheumatoid arthritis
Nature Reviews Rheumatology (2023)
-
Targeting PDK2 rescues stress-induced impaired brain energy metabolism
Molecular Psychiatry (2023)
-
Epigenomic profiling of glucocorticoid responses identifies cis-regulatory disruptions impacting steroid resistance in childhood acute lymphoblastic leukemia
Leukemia (2022)
-
Caspase-3/NLRP3 signaling in the mesenchymal stromal niche regulates myeloid-biased hematopoiesis
Stem Cell Research & Therapy (2021)