Abstract
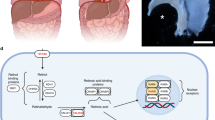
The diaphragm is an essential mammalian skeletal muscle, and defects in diaphragm development are the cause of congenital diaphragmatic hernias (CDHs), a common and often lethal birth defect. The diaphragm is derived from multiple embryonic sources, but how these give rise to the diaphragm is unknown, and, despite the identification of many CDH-associated genes, the etiology of CDH is incompletely understood. Using mouse genetics, we show that the pleuroperitoneal folds (PPFs), which are transient embryonic structures, are the source of the diaphragm's muscle connective tissue and regulate muscle development, and we show that the striking migration of PPF cells controls diaphragm morphogenesis. Furthermore, Gata4 mosaic mutations in PPF-derived muscle connective tissue fibroblasts result in the development of localized amuscular regions that are biomechanically weaker and more compliant, leading to CDH. Thus, the PPFs and muscle connective tissue are critical for diaphragm development, and mutations in PPF-derived fibroblasts are a source of CDH.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Perry, S.F., Similowski, T., Klein, W. & Codd, J.R. The evolutionary origin of the mammalian diaphragm. Respir. Physiol. Neurobiol. 171, 1–16 (2010).
Campbell, E.J.M., Agostoni, E. & Newsom Davis, J. The Respiratory Muscles: Mechanics and Neural Control (Lloyd-Luke, 1970).
Merrell, A.J. & Kardon, G. Development of the diaphragm—a skeletal muscle essential for mammalian respiration. FEBS J. 280, 4026–4035 (2013).
Raval, M.V., Wang, X., Reynolds, M. & Fischer, A.C. Costs of congenital diaphragmatic hernia repair in the United States—extracorporeal membrane oxygenation foots the bill. J. Pediatr. Surg. 46, 617–624 (2011).
Torfs, C.P., Curry, C.J., Bateson, T.F. & Honore, L.H. A population-based study of congenital diaphragmatic hernia. Teratology 46, 555–565 (1992).
Pober, B.R. Overview of epidemiology, genetics, birth defects, and chromosome abnormalities associated with CDH. Am. J. Med. Genet. C. Semin. Med. Genet. 145C, 158–171 (2007).
Pober, B.R. et al. Infants with Bochdalek diaphragmatic hernia: sibling precurrence and monozygotic twin discordance in a hospital-based malformation surveillance program. Am. J. Med. Genet. A. 138A, 81–88 (2005).
Holder, A.M. et al. Genetic factors in congenital diaphragmatic hernia. Am. J. Hum. Genet. 80, 825–845 (2007).
Veenma, D.C., de Klein, A. & Tibboel, D. Developmental and genetic aspects of congenital diaphragmatic hernia. Pediatr. Pulmonol. 47, 534–545 (2012).
Russell, M.K. et al. Congenital diaphragmatic hernia candidate genes derived from embryonic transcriptomes. Proc. Natl. Acad. Sci. USA 109, 2978–2983 (2012).
Arrington, C.B. et al. A family-based paradigm to identify candidate chromosomal regions for isolated congenital diaphragmatic hernia. Am. J. Med. Genet. A. 158A, 3137–3147 (2012).
Longoni, M. et al. Congenital diaphragmatic hernia interval on chromosome 8p23.1 characterized by genetics and protein interaction networks. Am. J. Med. Genet. A. 158A, 3148–3158 (2012).
Yu, L. et al. Variants in GATA4 are a rare cause of familial and sporadic congenital diaphragmatic hernia. Hum. Genet. 132, 285–292 (2013).
Kuo, C.T. et al. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. 11, 1048–1060 (1997).
Molkentin, J.D., Lin, Q., Duncan, S.A. & Olson, E.N. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. 11, 1061–1072 (1997).
Jay, P.Y. et al. Impaired mesenchymal cell function in Gata4 mutant mice leads to diaphragmatic hernias and primary lung defects. Dev. Biol. 301, 602–614 (2007).
Mendelsohn, C. et al. Function of the retinoic acid receptors (RARs) during development (II). Multiple abnormalities at various stages of organogenesis in RAR double mutants. Development 120, 2749–2771 (1994).
You, L.R. et al. Mouse lacking COUP-TFII as an animal model of Bochdalek-type congenital diaphragmatic hernia. Proc. Natl. Acad. Sci. USA 102, 16351–16356 (2005).
Ackerman, K.G. et al. Fog2 is required for normal diaphragm and lung development in mice and humans. PLoS Genet. 1, 58–65 (2005).
Coles, G.L. & Ackerman, K.G. Kif7 is required for the patterning and differentiation of the diaphragm in a model of syndromic congenital diaphragmatic hernia. Proc. Natl. Acad. Sci. USA 110, E1898–E1905 (2013).
Allan, D.W. & Greer, J.J. Embryogenesis of the phrenic nerve and diaphragm in the fetal rat. J. Comp. Neurol. 382, 459–468 (1997).
Babiuk, R.P., Zhang, W., Clugston, R., Allan, D.W. & Greer, J.J. Embryological origins and development of the rat diaphragm. J. Comp. Neurol. 455, 477–487 (2003).
Dietrich, S. et al. The role of SF/HGF and c-Met in the development of skeletal muscle. Development 126, 1621–1629 (1999).
Greer, J.J. et al. Structure of the primordial diaphragm and defects associated with nitrofen-induced CDH. J. Appl. Physiol. 89, 2123–2129 (2000).
Ackerman, K.G. & Greer, J.J. Development of the diaphragm and genetic mouse models of diaphragmatic defects. Am. J. Med. Genet. C. Semin. Med. Genet. 145C, 109–116 (2007).
Logan, M. et al. Expression of Cre recombinase in the developing mouse limb bud driven by a Prx1 enhancer. Genesis 33, 77–80 (2002).
Soriano, P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 21, 70–71 (1999).
Kardon, G., Harfe, B.D. & Tabin, C.J.A. Tcf4-positive mesodermal population provides a prepattern for vertebrate limb muscle patterning. Dev. Cell 5, 937–944 (2003).
Mathew, S.J. et al. Connective tissue fibroblasts and Tcf4 regulate myogenesis. Development 138, 371–384 (2011).
Clugston, R.D., Zhang, W. & Greer, J.J. Gene expression in the developing diaphragm: significance for congenital diaphragmatic hernia. Am. J. Physiol. Lung Cell. Mol. Physiol. 294, L665–L675 (2008).
Engleka, K.A. et al. Insertion of Cre into the Pax3 locus creates a new allele of Splotch and identifies unexpected Pax3 derivatives. Dev. Biol. 280, 396–406 (2005).
Vogan, K.J., Epstein, D.J., Trasler, D.G. & Gros, P. The splotch-delayed (Spd) mouse mutant carries a point mutation within the paired box of the Pax-3 gene. Genomics 17, 364–369 (1993).
Watt, A.J., Battle, M.A., Li, J. & Duncan, S.A. GATA4 is essential for formation of the proepicardium and regulates cardiogenesis. Proc. Natl. Acad. Sci. USA 101, 12573–12578 (2004).
Ackerman, K.G. et al. Congenital diaphragmatic defects: proposal for a new classification based on observations in 234 patients. Pediatr. Dev. Pathol. 15, 265–274 (2012).
Clugston, R.D. & Greer, J.J. Diaphragm development and congenital diaphragmatic hernia. Semin. Pediatr. Surg. 16, 94–100 (2007).
Maas, S.A., Ellis, B.J., Ateshian, G.A. & Weiss, J.A. FEBio: finite elements for biomechanics. J. Biomech. Eng. 134, 011005 (2012).
Strumpf, R.K., Humphrey, J.D. & Yin, F.C. Biaxial mechanical properties of passive and tetanized canine diaphragm. Am. J. Physiol. 265, H469–H475 (1993).
Keijzer, R., Liu, J., Deimling, J., Tibboel, D. & Post, M. Dual-hit hypothesis explains pulmonary hypoplasia in the nitrofen model of congenital diaphragmatic hernia. Am. J. Pathol. 156, 1299–1306 (2000).
Ackerman, K.G. et al. Gata4 is necessary for normal pulmonary lobar development. Am. J. Respir. Cell Mol. Biol. 36, 391–397 (2007).
Bladt, F., Riethmacher, D., Isenmann, S., Aguzzi, A. & Birchmeier, C. Essential role for the c-Met receptor in the migration of myogenic precursor cells into the limb bud. Nature 376, 768–771 (1995).
Maina, F. et al. Uncoupling of Grb2 from the Met receptor in vivo reveals complex roles in muscle development. Cell 87, 531–542 (1996).
Nakamura, K., Hongo, A., Kodama, J. & Hiramatsu, Y. The role of hepatocyte growth factor activator inhibitor (HAI)-1 and HAI-2 in endometrial cancer. Int. J. Cancer 128, 2613–2624 (2011).
Rojas, A. et al. GATA4 is a direct transcriptional activator of cyclin D2 and Cdk4 and is required for cardiomyocyte proliferation in anterior heart field–derived myocardium. Mol. Cell. Biol. 28, 5420–5431 (2008).
Yamak, A. et al. Cyclin D2 rescues size and function of GATA4 haplo-insufficient hearts. Am. J. Physiol. Heart Circ. Physiol. 303, H1057–H1066 (2012).
Domyan, E.T. et al. Roundabout receptors are critical for foregut separation from the body wall. Dev. Cell 24, 52–63 (2013).
Zhang, B. et al. Heparan sulfate deficiency disrupts developmental angiogenesis and causes congenital diaphragmatic hernia. J. Clin. Invest. 124, 209–221 (2014).
Veenma, D. et al. Copy number detection in discordant monozygotic twins of congenital diaphragmatic hernia (CDH) and esophageal atresia (EA) cohorts. Eur. J. Hum. Genet. 20, 298–304 (2012).
Wat, M.J. et al. Chromosome 8p23.1 deletions as a cause of complex congenital heart defects and diaphragmatic hernia. Am. J. Med. Genet. A. 149A, 1661–1677 (2009).
Kantarci, S. et al. Characterization of the chromosome 1q41q42.12 region, and the candidate gene DISP1, in patients with CDH. Am. J. Med. Genet. A. 152A, 2493–2504 (2010).
Veenma, D. et al. Comparable low-level mosaicism in affected and non affected tissue of a complex CDH patient. PLoS ONE 5, e15348 (2010).
Pu, W.T., Ishiwata, T., Juraszek, A.L., Ma, Q. & Izumo, S. GATA4 is a dosage-sensitive regulator of cardiac morphogenesis. Dev. Biol. 275, 235–244 (2004).
Wat, M.J. et al. Mouse model reveals the role of SOX7 in the development of congenital diaphragmatic hernia associated with recurrent deletions of 8p23.1. Hum. Mol. Genet. 21, 4115–4125 (2012).
Bosch, N. et al. Nucleotide, cytogenetic and expression impact of the human chromosome 8p23.1 inversion polymorphism. PLoS ONE 4, e8269 (2009).
Giglio, S. et al. Olfactory receptor-gene clusters, genomic-inversion polymorphisms, and common chromosome rearrangements. Am. J. Hum. Genet. 68, 874–883 (2001).
Reamon-Buettner, S.M. & Borlak, J. GATA4 zinc finger mutations as a molecular rationale for septation defects of the human heart. J. Med. Genet. 42, e32 (2005).
Reamon-Buettner, S.M., Cho, S.H. & Borlak, J. Mutations in the 3′-untranslated region of GATA4 as molecular hotspots for congenital heart disease (CHD). BMC Med. Genet. 8, 38 (2007).
Biesecker, L.G. & Spinner, N.B. A genomic view of mosaicism and human disease. Nat. Rev. Genet. 14, 307–320 (2013).
Tang, S.H., Silva, F.J., Tsark, W.M. & Mann, J.R.A. Cre/loxP-deleter transgenic line in mouse strain 129S1/SvImJ. Genesis 32, 199–202 (2002).
Muzumdar, M.D., Tasic, B., Miyamichi, K., Li, L. & Luo, L. A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605 (2007).
Wan, Y., Otsuna, H., Chien, C.B. & Hansen, C. An interactive visualization tool for multi-channel confocal microscopy data in neurobiology research. IEEE Trans. Vis. Comput. Graph. 15, 1489–1496 (2009).
Veronda, D.R. & Westmann, R.A. Mechanical characterization of skin-finite deformations. J. Biomech. 3, 111–124 (1970).
Mooney, M. A theory of large elastic deformation. J. Appl. Phys. 11, 582–592 (1940).
Acknowledgements
We thank C. Rodesch at the University of Utah Imaging Core for help with microscopy, S. Merchant for help with microCT imaging, Y. Wan and C. Hansen for Fluorender analysis, M. Hockin and M.R. Capecchi for Cre protein, and M. Colasanto, N. Elde, L.B. Jorde, A. Keefe, A. Letsou, L.C. Murtaugh and C.J. Tabin for critical comments on the manuscript. A.J.M. was supported by a University of Utah Graduate Fellowship, FEBio analysis is supported by US National Institutes of Health (NIH) grant R01GM083925 to J.A.W. and G.A. Ateshian, and Fluorender analysis is supported by US NIH grant R01GM098151 to C. Hansen. This research was supported by US NIH grant R01HD053728 and March of Dimes grant FY12-405 to G.K.
Author information
Authors and Affiliations
Contributions
A.J.M. conducted experiments, analyzed data and wrote the manuscript. Z.D.F. conducted two-photon experiments. B.J.E. and J.A.W. contributed finite element model analysis. J.A.L. managed the mouse colony. G.K. conducted experiments, analyzed data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 PPF cells migrate dynamically over the liver and septum transversum.
(a) Trajectories of individual GFP+ PPF cells labeled in E12.5 Prx1-creTg/+; RosamTmG/+ diaphragm explanted, cultured and imaged via two-photon microscopy for 4.6 h. Each gray arrow shows the absolute movement of each cell during 4.6 h, and each black arrow shows the movement of each cell normalized by the movement of the body wall. Arrows on the right were normalized by the average movement of the right body wall (arrow within the red circle), and arrows on the left were normalized by the average movement of the left body wall (arrow within the green circle). (b) Movement of individual cells during 4.6 h (boxed region in a). The line of dots shows the movement of each cell during the nine 30.7-min intervals. (a,b) Movement of the cells shown in Supplementary Video 2. The color-coded trajectories correspond to the color-coded cells in Supplementary Video 2. Scale bars, 500 μm (a), 125 μm (b).
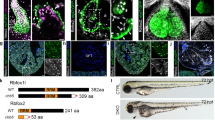
Supplementary Figure 2 Genetic labeling and ablation with the Prx1-cre transgene.
(a) The Prx1-cre transgene labels some cells in the lungs (n > 3/3). (b,c) Lung alveoli appear largely unaffected in mutant Prx1-creTg/+; Gata4Δ/fl mice. (d–i) The Prx1-cre transgene causes incomplete deletion of Gata4 in the PPFs at E12.5 (n = 4/4). Left insets show incomplete Prx1-cre–mediated recombination of Rosa26LacZ and Gata4fl, and right insets show more complete Prx1-cre–mediated recombination of Rosa26LacZ and Gata4fl. (a) Whole-mount β-galactosidase staining. (b–g) Section immunofluorescence. Scale bars, 1 mm (a), 50 μm (b,c), 100 μm (d–i), 20 μm (insets in d–i).
Supplementary Figure 3 Cell cycle proteins are downregulated in PPF cells with deletion of Gata4.
(a–l) Cdk4 (a–f) and cyclin D2 (g–l) are downregulated in Gata4-null PPF cells. E12.5 section immunofluorescence. Scale bars, 100 μm (a–l), 20 μm (insets in a–l).
Supplementary Figure 4 Gata4-null fibroblasts inhibit the proliferation of myogenic cells.
(a) When cultured, Gata4-null, as compared to wild-type, PPF fibroblasts proliferate less (n = 3 technical replicates). (b,c) When cultured for 48 h with Gata4-null PPF fibroblasts, as compared with Gata4-heterozygous fibroblasts, there are decreased cell numbers (b; n = 5 biological replicates, each with 3 technical replicates) and proliferation (c; n = 3 technical replicates) of Pax7+MyoD+ diaphragmatic myogenic cells. Micrographs show immunofluorescence labeling of fibroblasts (a) or myogenic cells (b,c). Scale bar, 50 μm. For charts, bars show mean values ± 1 s.e.m. P-values are from two-tailed Student’s t tests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4 and Supplementary Table 1. (PDF 1441 kb)
PPF cells actively migrate during diaphragm morphogenesis.
5.5-h time-lapse. GFP+ PPF cells at the leading edge migrate ventrally and medially (upper left to lower right in the video) across the surface of the liver in an explant of E12.5 Prx1-creTg/+; RosamTmG/+ diaphragm (with attached ribs and underlying liver). The location corresponding to the video is indicated by an asterisk in Figure 1c. The interval between frames is 5.6 min. The two red lines show the trajectory of the midpoint of the two labeled cells. Note that in frame 17 the entire diaphragm shifted down and at the end of the movie the diaphragm gradually sinks out of the field of view. Scale bar, 50 μm. (MOV 10814 kb)
PPF cells actively migrate across the liver surface during diaphragm morphogenesis.
4.6-h time-lapse of GFP+ PPF cells migrating ventrally across the surface of the liver in an explant of E12.5 Prx1-creTg/+; RosamTmG/+ diaphragm (with attached ribs and underlying liver). Ten individual fibroblasts are color-coded and tracked. The trajectories of selected cells are shown in Supplementary Figure 1. Scale bar, 500 μm. (MOV 5065 kb)
FEM modeling demonstrates that a hernia develops when the amuscular region is more compliant than the surrounding muscular region.
Top view. (MOV 231 kb)
FEM modeling demonstrates that a hernia develops when an amuscular region is more compliant than the surrounding muscular region.
Cross-sectional view. (MOV 116 kb)
Normal morphology of embryonic mouse lungs as shown by microCT.
Lateral view of the E18.5 lungs of control Prx1-creTg/+; Gata4fl/+ mice, with right lungs shown in blue. (MOV 7236 kb)
Lungs are malformed in regions associated with hernias as shown by microCT.
Lateral E18.5 lungs of mutant Prx1-creTg/+; Gata4fl/+ mice, with right lungs shown in blue. The divot in right lungs corresponds to the herniated region. (MOV 6200 kb)
Costal muscle progenitors normally develop within the pleuroperitoneal folds.
At E12.5, myogenic progenitors normally develop intermingled with and surrounded by PPF cells, and low levels of apoptotic cells are present in control Prx1-creTg/+; Gata4fl/+; RosamTmG/+ diaphragms. Three-dimensional view of whole-mount immunofluorescence of Pax7+MyoD+ muscle progenitors (red), GFP+ wild-type PPF cells (green) and TUNEL+ apoptotic cells (blue) at E12.5. Scale bar, 100 μm. (MOV 1641 kb)
Early increase in apoptosis and defects in localization of muscle progenitors lead to CDH.
At E12.5, there is a marked increase in apoptotic cells and a decrease in costal muscle progenitors, and muscle progenitors are largely absent from regions with Gata4-null PPF cells in Prx1-creTg/+; Gata4fl/+; RosamTmG/+ diaphragms. Three-dimensional view of whole-mount immunofluorescence of Pax7+MyoD+ muscle progenitors (red), GFP+ Gata4-null PPF cells (green) and TUNEL+ apoptotic cells (blue) at E12.5. Scale bar, 100 μm. (MOV 1420 kb)
Costal muscle progenitors normally proliferate within the pleuroperitoneal folds.
At E12.5, myogenic progenitors proliferate at high levels and are intermingled with and surrounded by PPF cells in control Prx1-creTg/+; Gata4fl/+; RosamTmG/+ diaphragms. Three-dimensional view of whole-mount immunofluorescence of Pax7+MyoD+ muscle progenitors (red), GFP+ PPF cells (green) and EdU+ proliferative cells (blue) at E12.5. Scale bar, 100 μm. (MOV 2203 kb)
Early defects in proliferation and localization of muscle progenitors lead to CDH.
At E12.5, there is a marked decrease in proliferating cells and a decrease in costal muscle progenitors, and muscle progenitors are largely absent from regions with Gata4-null PPF cells in Prx1-creTg/+; Gata4fl/+; RosamTmG/+ diaphragms. Three-dimensional view of whole-mount immunofluorescence of Pax7+MyoD+ muscle progenitors (red), GFP+ Gata4-null PPF cells (green) and EdU+ proliferative cells (blue) at E12.5. Scale bar, 100 μm. (MOV 1391 kb)
Rights and permissions
About this article
Cite this article
Merrell, A., Ellis, B., Fox, Z. et al. Muscle connective tissue controls development of the diaphragm and is a source of congenital diaphragmatic hernias. Nat Genet 47, 496–504 (2015). https://doi.org/10.1038/ng.3250
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3250
This article is cited by
-
Congenital diaphragmatic hernia
Nature Reviews Disease Primers (2022)
-
Origins, potency, and heterogeneity of skeletal muscle fibro-adipogenic progenitors—time for new definitions
Skeletal Muscle (2021)
-
Sudden onset dyspnea caused by Bochdalek diaphragmatic hernia in an adult: a case report
Journal of Medical Case Reports (2021)
-
Genetics of diaphragmatic hernia
European Journal of Human Genetics (2021)
-
Survival of infants with congenital diaphragmatic hernia in California: impact of hospital, clinical, and sociodemographic factors
Journal of Perinatology (2020)