Abstract
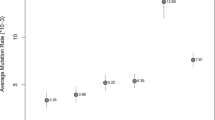
Mutations are the fundamental source of biological variation, and their rate is a crucial parameter for evolutionary and medical studies. Here we used whole-genome sequence data from 753 Icelandic males, grouped into 274 patrilines, to estimate the point mutation rate for 21.3 Mb of male-specific Y chromosome (MSY) sequence, on the basis of 1,365 meioses (47,123 years). The combined mutation rate for 15.2 Mb of X-degenerate (XDG), X-transposed (XTR) and ampliconic excluding palindromes (rAMP) sequence was 8.71 × 10−10 mutations per position per year (PPPY). We observed a lower rate (P = 0.04) of 7.37 × 10−10 PPPY for 6.1 Mb of sequence from palindromes (PAL), which was not statistically different from the rate of 7.2 × 10−10 PPPY for paternally transmitted autosomes1. We postulate that the difference between PAL and the other MSY regions may provide an indication of the rate at which nascent autosomal and PAL de novo mutations are repaired as a result of gene conversion.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Kong, A. et al. Rate of de novo mutations and the importance of father's age to disease risk. Nature 488, 471–475 (2012).
Conrad, D.F. et al. Variation in genome-wide mutation rates within and between human families. Nat. Genet. 43, 712–714 (2011).
Michaelson, J.J. et al. Whole-genome sequencing in autism identifies hot spots for de novo germline mutation. Cell 151, 1431–1442 (2012).
Roach, J.C. et al. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science 328, 636–639 (2010).
Sun, J.X. et al. A direct characterization of human mutation based on microsatellites. Nat. Genet. 44, 1161–1165 (2012).
Campbell, C.D. et al. Estimating the human mutation rate using autozygosity in a founder population. Nat. Genet. 44, 1277–1281 (2012).
Skaletsky, H. et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423, 825–837 (2003).
Hallast, P., Balaresque, P., Bowden, G.R., Ballereau, S. & Jobling, M.A. Recombination dynamics of a human Y-chromosomal palindrome: rapid GC-biased gene conversion, multi-kilobase conversion tracts, and rare inversions. PLoS Genet. 9, e1003666 (2013).
Rozen, S. et al. Abundant gene conversion between arms of palindromes in human and ape Y chromosomes. Nature 423, 873–876 (2003).
Heyer, E., Puymirat, J., Dieltjes, P., Bakker, E. & de Knijff, P. Estimating Y chromosome specific microsatellite mutation frequencies using deep rooting pedigrees. Hum. Mol. Genet. 6, 799–803 (1997).
Xue, Y. et al. Human Y chromosome base-substitution mutation rate measured by direct sequencing in a deep-rooting pedigree. Curr. Biol. 19, 1453–1457 (2009).
Poznik, G.D. et al. Sequencing Y chromosomes resolves discrepancy in time to common ancestor of males versus females. Science 341, 562–565 (2013).
Francalacci, P. et al. Low-pass DNA sequencing of 1200 Sardinians reconstructs European Y-chromosome phylogeny. Science 341, 565–569 (2013).
Scozzari, R. et al. An unbiased resource of novel SNP markers provides a new chronology for the human Y chromosome and reveals a deep phylogenetic structure in Africa. Genome Res. 24, 535–544 (2014).
Schrider, D.R., Hourmozdi, J.N. & Hahn, M.W. Pervasive multinucleotide mutational events in eukaryotes. Curr. Biol. 21, 1051–1054 (2011).
Ballantyne, K.N. et al. Mutability of Y-chromosomal microsatellites: rates, characteristics, molecular bases, and forensic implications. Am. J. Hum. Genet. 87, 341–353 (2010).
Cruciani, F. et al. A revised root for the human Y chromosomal phylogenetic tree: the origin of patrilineal diversity in Africa. Am. J. Hum. Genet. 88, 814–818 (2011).
Elhaik, E., Tatarinova, T.V., Klyosov, A.A. & Graur, D. The 'extremely ancient' chromosome that isn't: a forensic bioinformatic investigation of Albert Perry's X-degenerate portion of the Y chromosome. Eur. J. Hum. Genet. 22, 1111–1116 (2014).
Mendez, F.L. et al. An African American paternal lineage adds an extremely ancient root to the human Y chromosome phylogenetic tree. Am. J. Hum. Genet. 92, 454–459 (2013).
Scally, A. & Durbin, R. Revising the human mutation rate: implications for understanding human evolution. Nat. Rev. Genet. 13, 745–753 (2012).
Walsh, B. Estimating the time to the most recent common ancestor for the Y chromosome or mitochondrial DNA for a pair of individuals. Genetics 158, 897–912 (2001).
Behar, D.M. et al. A “Copernican” reassessment of the human mitochondrial DNA tree from its root. Am. J. Hum. Genet. 90, 675–684 (2012).
Ingman, M., Kaessmann, H., Paabo, S. & Gyllensten, U. Mitochondrial genome variation and the origin of modern humans. Nature 408, 708–713 (2000).
McLaren, W. et al. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics 26, 2069–2070 (2010).
Rosser, Z.H., Balaresque, P. & Jobling, M.A. Gene conversion between the X chromosome and the male-specific region of the Y chromosome at a translocation hotspot. Am. J. Hum. Genet. 85, 130–134 (2009).
Trombetta, B., Cruciani, F., Underhill, P.A., Sellitto, D. & Scozzari, R. Footprints of X-to-Y gene conversion in recent human evolution. Mol. Biol. Evol. 27, 714–725 (2010).
Trombetta, B., Sellitto, D., Scozzari, R. & Cruciani, F. Inter- and intraspecies phylogenetic analyses reveal extensive X-Y gene conversion in the evolution of gametologous sequences of human sex chromosomes. Mol. Biol. Evol. 31, 2108–2123 (2014).
Gudbjartsson, D.F. et al. Sequence variants from whole genome sequencing a large group of Icelanders. Sci. Data (in the press).
Martin, E.R. et al. SeqEM: an adaptive genotype-calling approach for next-generation sequencing studies. Bioinformatics 26, 2803–2810 (2010).
Harris, R.S. Improved Pairwise Alignment of Genomic DNA. PhD thesis. Penn. State Univ. (2007).
Repping, S. et al. High mutation rates have driven extensive structural polymorphism among human Y chromosomes. Nat. Genet. 38, 463–467 (2006).
Acknowledgements
We thank E. Haraldsdóttir for help with processing some of the whole-genome sequencing data and K.S.H. Moore for help with calling SNP microarray genotypes. A.W.E. was funded by a grant from Rannís, Icelandic Student Research Fund (1103340061). A.J. was funded by the EUROTAST Marie Curie Framework Programme 7 Initial Training Network (290344).
Author information
Authors and Affiliations
Contributions
A.H., A.W.E. and K.S. planned and directed the research. A.H. and A.W.E. analyzed the data, with A.K., V.B.G., E.D.G., A.J. and S.S.E. providing assistance with particular tasks. Á.S. performed the Sanger sequencing for the validation of de novo mutations. A.H., A.W.E. and K.S. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
A.H., V.B.G., A.S., E.D.G., A.K. and K.S. are employees of deCODE Genetics/Amgen.
Integrated supplementary information
Supplementary Figure 1 An example of a de novo mutation at paralogous positions.
We examine a G>A de novo mutation in the largest patriline with 86 meioses (the same patriline shown in Fig. 1), where sequence reads with the mutation map to three paralogous positions that are flanked by identical sequence in the human reference genome (NCBI b36), each shown in a separate depiction of the patriline: (a) 23,235,573 (rAMP7), (b) 24,071,677 (PAL1 arm 1), (c) 26,710,008 (PAL1 arm 2) and (d) with all reads combined (adjusting for the reverse complement orientation of reads at position 24,071,677 relative to positions 23,235,573 and 26,710,008). The allele state of the human reference sequence is indicated after the text "ref." Each square represents a male in the patriline, with vertical position scaled by birth year and the lines between squares representing Y-chromosome transmission events. The filled squares represent males demarcating the branches to which mutations can be assigned. Males with WGS data are indicated underneath a subset of filled squares by counts of alleles mapped to forward and reverse strands at the shown position(s) and inside each such square is the genotype called on the basis of these alleles. When examined separately, the genotypes called for the WGS males at the three positions provide inconsistent results about the branch on which the mutation occurred and the number of mutations required to account for the distribution of genotypes in the patriline. However, when the genotypes at all three positions are combined (d), it becomes clear that a single G>A mutation occurred on the branch labeled with the mutation event. We note, however, that it is not possible to resolve at which of the three positions the mutation occurred. Thus, for the purposes of this mutation rate study, each of the three paralogous positions was assigned one-third of a mutation, i.e., a weight of 1/3.
Supplementary Figure 2 The XDG normalized sequence depth of branches by sequence region and haplogroup.
Results are shown for the 482 branches with an average XDG sequence depth >10×. For each branch, the average depth in each of 27 regions of the 4 sequence classes was normalized through division by its average XDG sequence depth. This was done because the branches have very different baseline expectations of sequence depth, which depend on the number of males who belong to them and the amount of sequencing undertaken for each. Normalization by average XDG sequence depth allows meaningful comparison of sequence depth among branches. Moreover, as the XDG region is mostly present in single copy, the normalized value provides some information about the relative magnitude of change in copy number. For each sequence region and haplogroup, we then calculated and plotted the average XDG normalized sequence depth with 95% confidence intervals. The greatest differences are seen for region PAL_IR2. Here we see that males belonging to haplogroup Q1a3a appear to have just a single copy of each orientation of IR2 (around 70 and 80 kb in length, respectively), as seen in the NCBI Build 36 reference sequence, whereas males from the other haplogroups have an excess of 17–30% of reads that map to these small sequence regions, indicating a greater copy number of at least part of the IR2 sequence. Other regions that exhibit differences between haplogroups are rAMP2, where the XDG normalized sequence depth is greater in E1b1 and Q1a3a than in the other haplogroups, and XTR2, where R1b1a and Q1a3a exhibit greater XDG normalized sequence depth than the rest. For most other sequence regions, there is little evidence for differences in copy number among haplogroups. On the basis of this evidence, we deem it unlikely that our mutation rate estimates are affected by copy number variation of MSY sequence.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1 and 2, Supplementary Tables 1, 5–7, 10 and 11, and Supplementary Note. (PDF 656 kb)
Supplementary Table 2
Summary of 739 branches in 274 patrilines with weighted sums of mutations. (XLSX 98 kb)
Supplementary Table 3
Summary of the 274 patrilines used in the study. (XLSX 13 kb)
Supplementary Table 4
Full list of 1,456 candidate de novo mutations at 2,050 positions with weights. (XLSX 258 kb)
Supplementary Table 8
Comparison of WGS and Illumina SNP chip genotypes. (XLSX 34 kb)
Supplementary Table 9
Mutation rate by haplogroup and sequence region for branches with XDG sequence depth >10×. (XLSX 18 kb)
Rights and permissions
About this article
Cite this article
Helgason, A., Einarsson, A., Guðmundsdóttir, V. et al. The Y-chromosome point mutation rate in humans. Nat Genet 47, 453–457 (2015). https://doi.org/10.1038/ng.3171
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3171
This article is cited by
-
Characterizing the evolution and phenotypic impact of ampliconic Y chromosome regions
Nature Communications (2023)
-
Origin and diffusion of human Y chromosome haplogroup J1-M267
Scientific Reports (2021)
-
The paternal and maternal genetic history of Vietnamese populations
European Journal of Human Genetics (2020)
-
Cultural variation impacts paternal and maternal genetic lineages of the Hmong-Mien and Sino-Tibetan groups from Thailand
European Journal of Human Genetics (2020)
-
Mosaic loss of human Y chromosome: what, how and why
Human Genetics (2020)