Abstract
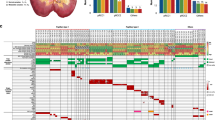
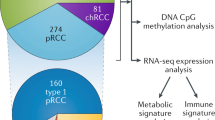
To further understand the molecular distinctions between kidney cancer subtypes, we analyzed exome, transcriptome and copy number alteration data from 167 primary human tumors that included renal oncocytomas and non–clear cell renal cell carcinomas (nccRCCs), consisting of papillary (pRCC), chromophobe (chRCC) and translocation (tRCC) subtypes. We identified ten significantly mutated genes in pRCC, including MET, NF2, SLC5A3, PNKD and CPQ. MET mutations occurred in 15% (10/65) of pRCC samples and included previously unreported recurrent activating mutations. In chRCC, we found TP53, PTEN, FAAH2, PDHB, PDXDC1 and ZNF765 to be significantly mutated. Gene expression analysis identified a five-gene set that enabled the molecular classification of chRCC, renal oncocytoma and pRCC. Using RNA sequencing, we identified previously unreported gene fusions, including ACTG1-MITF fusion. Ectopic expression of the ACTG1-MITF fusion led to cellular transformation and induced the expression of downstream target genes. Finally, we observed upregulation of the anti-apoptotic factor BIRC7 in MiTF-high RCC tumors, suggesting a potential therapeutic role for BIRC7 inhibitors.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Siegel, R., Naishadham, D. & Jemal, A. Cancer statistics, 2013. CA Cancer J. Clin. 63, 11–30 (2013).
Peña-Llopis, S. et al. BAP1 loss defines a new class of renal cell carcinoma. Nat. Genet. 44, 751–759 (2012).
Sato, Y. et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat. Genet. 45, 860–867 (2013).
Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 499, 43–49 (2013).
Srigley, J.R. et al. The International Society of Urological Pathology (ISUP) Vancouver Classification of Renal Neoplasia. Am. J. Surg. Pathol. 37, 1469–1489 (2013).
Yusenko, M.V. Molecular pathology of renal oncocytoma: a review. Int. J. Urol. 17, 602–612 (2010).
Amin, M.B. et al. Chromophobe renal cell carcinoma: histomorphologic characteristics and evaluation of conventional pathologic prognostic parameters in 145 cases. Am. J. Surg. Pathol. 32, 1822–1834 (2008).
Eble, J.N., Sauter, G., Epstein, J.I. & Sesterhenn, I.A. Pathology and Genetics of Tumours of the Urinary System and Male Genital Organs (IARCPress, Lyon, France, 2004).
Schieda, N., Al-Subhi, M., Flood, T.A., El-Khodary, M. & McInnes, M.D. Diagnostic accuracy of segmental enhancement inversion for the diagnosis of renal oncocytoma using biphasic computed tomography (CT) and multiphase contrast-enhanced magnetic resonance imaging (MRI). Eur. Radiol. 24, 2787–2794 (2014).
Vargas, H.A. et al. Renal cortical tumors: use of multiphasic contrast-enhanced MR imaging to differentiate benign and malignant histologic subtypes. Radiology 264, 779–788 (2012).
Rosenkrantz, A.B. et al. MRI features of renal oncocytoma and chromophobe renal cell carcinoma. AJR Am. J. Roentgenol. 195, W421–W427 (2010).
Young, A.N. Editorial comment from Dr Young to Molecular pathology of renal oncocytoma: a review. Int. J. Urol. 17, 612–613 (2010).
Picken, M.M. Editorial comment from Dr Picken to Molecular pathology of renal oncocytoma: a review. Int. J. Urol. 17, 613–614 (2010).
Yusenko, M.V. Molecular pathology of chromophobe renal cell carcinoma: a review. Int. J. Urol. 17, 592–600 (2010).
Osunkoya, A.O. Editorial comment to Molecular pathology of chromophobe renal cell carcinoma: a review. Int. J. Urol. 17, 600–601 (2010).
Macher-Goeppinger, S. et al. Molecular heterogeneity of TFE3 activation in renal cell carcinomas. Mod. Pathol. 25, 308–315 (2012).
Bellmunt, J. & Dutcher, J. Targeted therapies and the treatment of non–clear cell renal cell carcinoma. Ann. Oncol. 24, 1730–1740 (2013).
Hagenkord, J.M., Gatalica, Z., Jonasch, E. & Monzon, F.A. Clinical genomics of renal epithelial tumors. Cancer Genet. 204, 285–297 (2011).
Linehan, W.M. & Ricketts, C.J. The metabolic basis of kidney cancer. Semin. Cancer Biol. 23, 46–55 (2013).
Popova, T. et al. Germline BAP1 mutations predispose to renal cell carcinomas. Am. J. Hum. Genet. 92, 974–980 (2013).
Farley, M.N. et al. A novel germline mutation in BAP1 predisposes to familial clear-cell renal cell carcinoma. Mol. Cancer Res. 11, 1061–1071 (2013).
Ricketts, C.J. et al. Succinate dehydrogenase kidney cancer: an aggressive example of the Warburg effect in cancer. J. Urol. 188, 2063–2071 (2012).
Schmidt, L. et al. Germline and somatic mutations in the tyrosine kinase domain of the MET proto-oncogene in papillary renal carcinomas. Nat. Genet. 16, 68–73 (1997).
Schmidt, L. et al. Novel mutations of the MET proto-oncogene in papillary renal carcinomas. Oncogene 18, 2343–2350 (1999).
Argani, P. et al. Xp11 translocation renal cell carcinoma in adults: expanded clinical, pathologic, and genetic spectrum. Am. J. Surg. Pathol. 31, 1149–1160 (2007).
Peña-Llopis, S. & Brugarolas, J. Simultaneous isolation of high-quality DNA, RNA, miRNA and proteins from tissues for genomic applications. Nat. Protoc. 8, 2240–2255 (2013).
Forbes, S.A. et al. COSMIC (the Catalogue of Somatic Mutations in Cancer): a resource to investigate acquired mutations in human cancer. Nucleic Acids Res. 38, D652–D657 (2010).
Nik-Zainal, S. et al. Mutational processes molding the genomes of 21 breast cancers. Cell 149, 979–993 (2012).
Pfeifer, G.P. Mutagenesis at methylated CpG sequences. Curr. Top. Microbiol. Immunol. 301, 259–281 (2006).
Ng, P.C. & Henikoff, S. Accounting for human polymorphisms predicted to affect protein function. Genome Res. 12, 436–446 (2002).
Ramensky, V., Bork, P. & Sunyaev, S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 30, 3894–3900 (2002).
González-Pérez, A. & López-Bigas, N. Improving the assessment of the outcome of nonsynonymous SNVs with a consensus deleteriousness score, Condel. Am. J. Hum. Genet. 88, 440–449 (2011).
Kan, Z. et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature 466, 869–873 (2010).
Wang, W. et al. Structural characterization of autoinhibited c-Met kinase produced by coexpression in bacteria with phosphatase. Proc. Natl. Acad. Sci. USA 103, 3563–3568 (2006).
Miller, M. et al. Structural basis of oncogenic activation caused by point mutations in the kinase domain of the MET proto-oncogene: modeling studies. Proteins 44, 32–43 (2001).
Gandino, L., Longati, P., Medico, E., Prat, M. & Comoglio, P.M. Phosphorylation of serine 985 negatively regulates the hepatocyte growth factor receptor kinase. J. Biol. Chem. 269, 1815–1820 (1994).
Toker, L. et al. Inositol-related gene knockouts mimic lithium's effect on mitochondrial function. Neuropsychopharmacology 39, 319–328 (2014).
Patel, M.S., Nemeria, N.S., Furey, W. & Jordan, F. The pyruvate dehydrogenase complexes: structure-based function and regulation. J. Biol. Chem. 289, 16615–16623 (2014).
Imbard, A. et al. Molecular characterization of 82 patients with pyruvate dehydrogenase complex deficiency. Structural implications of novel amino acid substitutions in E1 protein. Mol. Genet. Metab. 104, 507–516 (2011).
Quintana, E. et al. PDH E1β deficiency with novel mutations in two patients with Leigh syndrome. J. Inherit. Metab. Dis. 32, 339–343 (2009).
Linehan, W.M., Srinivasan, R. & Schmidt, L.S. The genetic basis of kidney cancer: a metabolic disease. Nat. Rev. Urol. 7, 277–285 (2010).
Steinberg, G.R. & Kemp, B.E. AMPK in health and disease. Physiol. Rev. 89, 1025–1078 (2009).
Scott, J.W., Ross, F.A., Liu, J.K. & Hardie, D.G. Regulation of AMP-activated protein kinase by a pseudosubstrate sequence on the γ subunit. EMBO J. 26, 806–815 (2007).
Davis, C.F. et al. The somatic genomic landscape of chromophobe renal cell carcinoma. Cancer Cell 26, 319–330 (2014).
Burwinkel, B. et al. Fatal congenital heart glycogenosis caused by a recurrent activating R531Q mutation in the γ2-subunit of AMP-activated protein kinase (PRKAG2), not by phosphorylase kinase deficiency. Am. J. Hum. Genet. 76, 1034–1049 (2005).
Carling, D., Mayer, F.V., Sanders, M.J. & Gamblin, S.J. AMP-activated protein kinase: nature's energy sensor. Nat. Chem. Biol. 7, 512–518 (2011).
Davies, J.K. et al. Characterization of the role of γ2 R531G mutation in AMP-activated protein kinase in cardiac hypertrophy and Wolff-Parkinson-White syndrome. Am. J. Physiol. Heart Circ. Physiol. 290, H1942–H1951 (2006).
de Moor, R.A. et al. Hepatocellular carcinoma in glycogen storage disease type IV. Arch. Dis. Child. 82, 479–480 (2000).
Manzia, T.M. et al. Glycogen storage disease type Ia and VI associated with hepatocellular carcinoma: two case reports. Transplant. Proc. 43, 1181–1183 (2011).
Calderaro, J. et al. Molecular characterization of hepatocellular adenomas developed in patients with glycogen storage disease type I. J. Hepatol. 58, 350–357 (2013).
Fu, L., Wang, G., Shevchuk, M.M., Nanus, D.M. & Gudas, L.J. Generation of a mouse model of von Hippel–Lindau kidney disease leading to renal cancers by expression of a constitutively active mutant of HIF1. Cancer Res. 71, 6848–6856 (2011).
Jeon, S.M., Chandel, N.S. & Hay, N. AMPK regulates NADPH homeostasis to promote tumour cell survival during energy stress. Nature 485, 661–665 (2012).
Milacic, M. et al. Annotating cancer variants and anti-cancer therapeutics in reactome. Cancers (Basel) 4, 1180–1211 (2012).
Croft, D. et al. Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res. 39, D691–D697 (2011).
Nagashima, Y., Kuroda, N. & Yao, M. Transition of organizational category on renal cancer. Jpn. J. Clin. Oncol. 43, 233–242 (2013).
Beleut, M. et al. Integrative genome-wide expression profiling identifies three distinct molecular subgroups of renal cell carcinoma with different patient outcome. BMC Cancer 12, 310 (2012).
Rohan, S. et al. Gene expression profiling separates chromophobe renal cell carcinoma from oncocytoma and identifies vesicular transport and cell junction proteins as differentially expressed genes. Clin. Cancer Res. 12, 6937–6945 (2006).
Venkateswarlu, K. & Cullen, P.J. Molecular cloning and functional characterization of a human homologue of centaurin-α. Biochem. Biophys. Res. Commun. 262, 237–244 (1999).
Zimmermann, P. The prevalence and significance of PDZ domain–phosphoinositide interactions. Biochim. Biophys. Acta 1761, 947–956 (2006).
Weimer, J.M., Chattopadhyay, S., Custer, A.W. & Pearce, D.A. Elevation of Hook1 in a disease model of Batten disease does not affect a novel interaction between ankyrin G and Hook1. Biochem. Biophys. Res. Commun. 330, 1176–1181 (2005).
Palmer, R.E. et al. Induction of BAIAP3 by the EWS-WT1 chimeric fusion implicates regulated exocytosis in tumorigenesis. Cancer Cell 2, 497–505 (2002).
Cheng, H., Fukushima, T., Takahashi, N., Tanaka, H. & Kataoka, H. Hepatocyte growth factor activator inhibitor type 1 regulates epithelial to mesenchymal transition through membrane-bound serine proteinases. Cancer Res. 69, 1828–1835 (2009).
Yusenko, M.V. et al. High-resolution DNA copy number and gene expression analyses distinguish chromophobe renal cell carcinomas and renal oncocytomas. BMC Cancer 9, 152 (2009).
Righi, L., Rapa, I., Votta, A., Papotti, M. & Sapino, A. Human achaete-scute homolog-1 expression in neuroendocrine breast carcinoma. Virchows Arch. 460, 415–421 (2012).
Palmieri, F. The mitochondrial transporter family SLC25: identification, properties and physiopathology. Mol. Aspects Med. 34, 465–484 (2013).
Monzon, F.A. et al. Whole genome SNP arrays as a potential diagnostic tool for the detection of characteristic chromosomal aberrations in renal epithelial tumors. Mod. Pathol. 21, 599–608 (2008).
Choueiri, T.K. et al. Phase II and biomarker study of the dual MET/VEGFR2 inhibitor foretinib in patients with papillary renal cell carcinoma. J. Clin. Oncol. 31, 181–186 (2013).
Lager, D.J., Huston, B.J., Timmerman, T.G. & Bonsib, S.M. Papillary renal tumors. Morphologic, cytochemical, and genotypic features. Cancer 76, 669–673 (1995).
Steingrímsson, E., Copeland, N.G. & Jenkins, N.A. Melanocytes and the microphthalmia transcription factor network. Annu. Rev. Genet. 38, 365–411 (2004).
Haq, R. & Fisher, D.E. Biology and clinical relevance of the micropthalmia family of transcription factors in human cancer. J. Clin. Oncol. 29, 3474–3482 (2011).
Malouf, G.G. et al. Next-generation sequencing of translocation renal cell carcinoma reveals novel RNA splicing partners and frequent mutations of chromatin-remodeling genes. Clin. Cancer Res. 20, 4129–4140 (2014).
Maruyama, K. et al. Strawberry notch homologue 2 regulates osteoclast fusion by enhancing the expression of DC-STAMP. J. Exp. Med. 210, 1947–1960 (2013).
Dynek, J.N. et al. Microphthalmia-associated transcription factor is a critical transcriptional regulator of melanoma inhibitor of apoptosis in melanomas. Cancer Res. 68, 3124–3132 (2008).
Hoek, K.S. et al. Novel MITF targets identified using a two-step DNA microarray strategy. Pigment Cell Melanoma Res. 21, 665–676 (2008).
Yokoyama, S. et al. A novel recurrent mutation in MITF predisposes to familial and sporadic melanoma. Nature 480, 99–103 (2011).
Hemesath, T.J. et al. Microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev. 8, 2770–2780 (1994).
Hakimi, A.A., Pham, C.G. & Hsieh, J.J. A clear picture of renal cell carcinoma. Nat. Genet. 45, 849–850 (2013).
Gad, S. et al. Mutations in BHD and TP53 genes, but not in HNF1β gene, in a large series of sporadic chromophobe renal cell carcinoma. Br. J. Cancer 96, 336–340 (2007).
Contractor, H., Zariwala, M., Bugert, P., Zeisler, J. & Kovacs, G. Mutation of the p53 tumour suppressor gene occurs preferentially in the chromophobe type of renal cell tumour. J. Pathol. 181, 136–139 (1997).
Morgan, M. et al. ShortRead: a bioconductor package for input, quality assessment and exploration of high-throughput sequence data. Bioinformatics 25, 2607–2608 (2009).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
DePristo, M.A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011).
Saunders, C.T. et al. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics 28, 1811–1817 (2012).
Sherry, S.T. et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 29, 308–311 (2001).
Fu, W. et al. Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature 493, 216–220 (2013).
Seshagiri, S. et al. Recurrent R-spondin fusions in colon cancer. Nature 488, 660–664 (2012).
Alexandrov, L.B., Nik-Zainal, S., Wedge, D.C., Campbell, P.J. & Stratton, M.R. Deciphering signatures of mutational processes operative in human cancer. Cell Rep. 3, 246–259 (2013).
Brunet, J.P., Tamayo, P., Golub, T.R. & Mesirov, J.P. Metagenes and molecular pattern discovery using matrix factorization. Proc. Natl. Acad. Sci. USA 101, 4164–4169 (2004).
Dees, N.D. MuSiC: identifying mutational significance in cancer genomes. Genome Res. 22, 1589–1598 (2012).
Rudin, C.M. et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat. Genet. 44, 1111–1116 (2012).
Lawrence, M.S. et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218 (2013).
Wu, T.D. & Nacu, S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26, 873–881 (2010).
Robinson, M.D., McCarthy, D.J. & Smyth, G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Anders, S. & Huber, W. Differential expression analysis for sequence count data. Genome Biol. 11, R106 (2010).
Greenman, C.D. et al. PICNIC: an algorithm to predict absolute allelic copy number variation with microarray cancer data. Biostatistics 11, 164–175 (2010).
Tibshirani, R. & Wang, P. Spatial smoothing and hot spot detection for CGH data using the fused lasso. Biostatistics 9, 18–29 (2008).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550 (2005).
Jaiswal, B.S. et al. Somatic mutations in p85α promote tumorigenesis through class IA PI3K activation. Cancer Cell 16, 463–474 (2009).
Acknowledgements
The authors would like to acknowledge the Genentech DNA Sequencing, Oligo and Bioinformatics groups for their help with the project. We thank D. Bhatt, R. Bourgon, Z. Zhang, C. Klijn, M. Brauer and L. Johnson for their support during the course of this project. We would like to acknowledge K. Mukhyala for assistance in gene annotation. This work was supported in part by grants 1R01CA175754 (US National Institutes of Health (NIH)) and RP130603 (CPRIT; Texas, USA) to J.B. and grant 5R01CA154475-04 (US NIH) to I.P. Sample collection was supported in part by grant 5P30CA142543 (US NIH). J.B. is a Virginia Murchison Linthicum Endowed Scholar in medical research.
Author information
Authors and Affiliations
Contributions
S.D. and E.W.S. performed the exome and RNA-seq analyses. T.T.N. performed the simulation analysis. E.W.S. and J.S.G. conducted the mutational signature studies. S.D. and P.M.H. performed copy number analysis. A.P.-J., V.T.-T., E.H., Z.M., H.M.H. and S.P.-L. were responsible for sample collection, annotation, processing, and DNA and RNA extraction. Z.M. oversaw the collection of the various data types. Z.M. and Y.-J.C. performed validation of the fusions. P.K. facilitated sample procurement, selected samples for DNA and RNA extraction, oversaw FISH analyses and served as the pathologist for the study. J.R. and G.P. processed the RNA-seq reads. T.D.W. provided support for gene fusion prediction. K.T., C.H., C.J.H. and C.S.R. prepared the sequencing libraries. N.Z., K.B.P., S.C. and B.S.J. performed biological validation studies. S. Saleem collected information about the cases. V.M., Y.L., A.S. and I.P. facilitated sample procurement. L.N.K. and N.V.G. performed in silico analyses. J.G., V.J. and J.S. collected sequencing data. J.G. performed mutation validation. B.C. analyzed targeted capture data. S.H. and M.J. performed Sanger sequencing to validate indels. W.W. predicted the structural consequences of MET mutations. F.J.d.S. provided organizational support. J.B. and S. Seshagiri conceived the study and designed the experiments. E.W.S., S.D., Z.M., P.K., J.B. and S. Seshagiri wrote the manuscript, which was reviewed and edited by the other coauthors.
Corresponding authors
Ethics declarations
Competing interests
All Genentech-affiliated authors (S.D., E.W.S., Z.M., B.S.J., N.Z., T.T.N., K.B.P., Y.-J.C., S.C., S.H., M.J., J.G., K.T., C.H., C.J.H., J.S., C.S.R., V.J., W.W., P.M.H., B.C., J.S.G., J.R., G.P., T.D.W., F.J.d.S. and S. Seshagiri) hold Roche shares.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–29. (PDF 12430 kb)
Supplementary Tables 1–13
Supplementary Tables 1–13. (XLSX 1495 kb)
Source data
Rights and permissions
About this article
Cite this article
Durinck, S., Stawiski, E., Pavía-Jiménez, A. et al. Spectrum of diverse genomic alterations define non–clear cell renal carcinoma subtypes. Nat Genet 47, 13–21 (2015). https://doi.org/10.1038/ng.3146
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3146
This article is cited by
-
Metabolic alterations in hereditary and sporadic renal cell carcinoma
Nature Reviews Nephrology (2024)
-
Biphasic papillary (biphasic squamoid alveolar) renal cell carcinoma: a clinicopathologic and molecular study of 17 renal cell carcinomas including 10 papillary adenomas
Virchows Archiv (2024)
-
Evaluation of an institutional series of low-grade oncocytic tumor (LOT) of the kidney and review of the mutational landscape of LOT
Virchows Archiv (2023)
-
Crosstalk of organelles in Parkinson’s disease – MiT family transcription factors as central players in signaling pathways connecting mitochondria and lysosomes
Molecular Neurodegeneration (2022)
-
Inhibition of HSP 90 is associated with potent anti-tumor activity in Papillary Renal Cell Carcinoma
Journal of Experimental & Clinical Cancer Research (2022)