Abstract
Diffuse large B cell lymphoma (DLBCL) is the most common lymphoma subtype and is clinically aggressive. To identify genetic susceptibility loci for DLBCL, we conducted a meta-analysis of 3 new genome-wide association studies (GWAS) and 1 previous scan, totaling 3,857 cases and 7,666 controls of European ancestry, with additional genotyping of 9 promising SNPs in 1,359 cases and 4,557 controls. In our multi-stage analysis, five independent SNPs in four loci achieved genome-wide significance marked by rs116446171 at 6p25.3 (EXOC2; P = 2.33 × 10−21), rs2523607 at 6p21.33 (HLA-B; P = 2.40 × 10−10), rs79480871 at 2p23.3 (NCOA1; P = 4.23 × 10−8) and two independent SNPs, rs13255292 and rs4733601, at 8q24.21 (PVT1; P = 9.98 × 10−13 and 3.63 × 10−11, respectively). These data provide substantial new evidence for genetic susceptibility to this B cell malignancy and point to pathways involved in immune recognition and immune function in the pathogenesis of DLBCL.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
Accession codes
References
Siegel, R., Naishadham, D. & Jemal, A. Cancer statistics, 2013. CA Cancer J. Clin. 63, 11–30 (2013).
Flowers, C.R., Sinha, R. & Vose, J.M. Improving outcomes for patients with diffuse large B-cell lymphoma. CA Cancer J. Clin. 60, 393–408 (2010).
Wang, S.S. et al. Family history of hematopoietic malignancies and risk of non-Hodgkin lymphoma (NHL): a pooled analysis of 10,211 cases and 11,905 controls from the International Lymphoma Epidemiology Consortium (InterLymph). Blood 109, 3479–3488 (2007).
Goldin, L.R., Bjorkholm, M., Kristinsson, S.Y., Turesson, I. & Landgren, O. Highly increased familial risks for specific lymphoma subtypes. Br. J. Haematol. 146, 91–94 (2009).
Skibola, C.F. et al. Tumor necrosis factor (TNF) and lymphotoxin-α (LTA) polymorphisms and risk of non-Hodgkin lymphoma in the InterLymph Consortium. Am. J. Epidemiol. 171, 267–276 (2010).
Skibola, C.F. et al. Genetic variants at 6p21.33 are associated with susceptibility to follicular lymphoma. Nat. Genet. 41, 873–875 (2009).
Conde, L. et al. Genome-wide association study of follicular lymphoma identifies a risk locus at 6p21.32. Nat. Genet. 42, 661–664 (2010).
Smedby, K.E. et al. GWAS of follicular lymphoma reveals allelic heterogeneity at 6p21.32 and suggests shared genetic susceptibility with diffuse large B-cell lymphoma. PLoS Genet. 7, e1001378 (2011).
Vijai, J. et al. Susceptibility loci associated with specific and shared subtypes of lymphoid malignancies. PLoS Genet. 9, e1003220 (2013).
Tan, D.E. et al. Genome-wide association study of B cell non-Hodgkin lymphoma identifies 3q27 as a susceptibility locus in the Chinese population. Nat. Genet. 45, 804–807 (2013).
Wang, Z. et al. Improved imputation of common and uncommon SNPs with a new reference set. Nat. Genet. 44, 6–7 (2012).
Abecasis, G.R. et al. A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073 (2010).
Howie, B.N., Donnelly, P. & Marchini, J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 5, e1000529 (2009).
Wang, S.S. et al. Human leukocyte antigen class I and II alleles in non-Hodgkin lymphoma etiology. Blood 115, 4820–4823 (2010).
Ward, L.D. & Kellis, M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 40, D930–D934 (2012).
Barenboim, M. & Manke, T. ChroMoS: an integrated web tool for SNP classification, prioritization and functional interpretation. Bioinformatics 29, 2197–2198 (2013).
Ernst, J. et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473, 43–49 (2011).
Mukerji, J., Olivieri, K.C., Misra, V., Agopian, K.A. & Gabuzda, D. Proteomic analysis of HIV-1 Nef cellular binding partners reveals a role for exocyst complex proteins in mediating enhancement of intercellular nanotube formation. Retrovirology 9, 33 (2012).
Mantovani, A. & Balkwill, F. RalB signaling: a bridge between inflammation and cancer. Cell 127, 42–44 (2006).
Bodemann, B.O. & White, M.A. Ral GTPases and cancer: linchpin support of the tumorigenic platform. Nat. Rev. Cancer 8, 133–140 (2008).
Issaq, S.H., Lim, K.H. & Counter, C.M. Sec5 and Exo84 foster oncogenic Ras-mediated tumorigenesis. Mol. Cancer Res. 8, 223–231 (2010).
Kashatus, D.F. Ral GTPases in tumorigenesis: emerging from the shadows. Exp. Cell Res. 319, 2337–2342 (2013).
Di Bernardo, M.C. et al. A genome-wide association study identifies six susceptibility loci for chronic lymphocytic leukemia. Nat. Genet. 40, 1204–1210 (2008).
Berndt, S.I. et al. Genome-wide association study identifies multiple risk loci for chronic lymphocytic leukemia. Nat. Genet. 45, 868–876 (2013).
Wang, S.S. et al. Common gene variants in the tumor necrosis factor (TNF) and TNF receptor superfamilies and NF-κB transcription factors and non-Hodgkin lymphoma risk. PLoS ONE 4, e5360 (2009).
Wacholder, S., Yeager, M. & Liao, L.M. Invited commentary: more surprises from a gene desert. Am. J. Epidemiol. 175, 488–491 (2012).
Crowther-Swanepoel, D. et al. Common variants at 2q37.3, 8q24.21, 15q21.3 and 16q24.1 influence chronic lymphocytic leukemia risk. Nat. Genet. 42, 132–136 (2010).
Enciso-Mora, V. et al. A genome-wide association study of Hodgkin's lymphoma identifies new susceptibility loci at 2p16.1 (REL), 8q24.21 and 10p14 (GATA3). Nat. Genet. 42, 1126–1130 (2010).
Graham, M. & Adams, J.M. Chromosome 8 breakpoint far 3′ of the c-myc oncogene in a Burkitt's lymphoma 2;8 variant translocation is equivalent to the murine pvt-1 locus. EMBO J. 5, 2845–2851 (1986).
Love, C. et al. The genetic landscape of mutations in Burkitt lymphoma. Nat. Genet. 44, 1321–1325 (2012).
Savage, K.J. et al. MYC gene rearrangements are associated with a poor prognosis in diffuse large B-cell lymphoma patients treated with R-CHOP chemotherapy. Blood 114, 3533–3537 (2009).
Pasqualucci, L. et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat. Genet. 43, 830–837 (2011).
Oñate, S.A., Tsai, S.Y., Tsai, M.J. & O'Malley, B.W. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science 270, 1354–1357 (1995).
Novokhatska, O. et al. Adaptor proteins intersectin 1 and 2 bind similar proline-rich ligands but are differentially recognized by SH2 domain–containing proteins. PLoS ONE 8, e70546 (2013).
McGavin, M.K. et al. The intersectin 2 adaptor links Wiskott Aldrich Syndrome protein (WASp)-mediated actin polymerization to T cell antigen receptor endocytosis. J. Exp. Med. 194, 1777–1787 (2001).
Jia, X. et al. Imputing amino acid polymorphisms in human leukocyte antigens. PLoS ONE 8, e64683 (2013).
Howell, W.M. HLA and disease: guilt by association. Int. J. Immunogenet. 41, 1–12 (2014).
Klitz, W., Aldrich, C.L., Fildes, N., Horning, S.J. & Begovich, A.B. Localization of predisposition to Hodgkin disease in the HLA class II region. Am. J. Hum. Genet. 54, 497–505 (1994).
Price, P. et al. The genetic basis for the association of the 8.1 ancestral haplotype (A1, B8, DR3) with multiple immunopathological diseases. Immunol. Rev. 167, 257–274 (1999).
Kumar, V. et al. Common variants on 14q32 and 13q12 are associated with DLBCL susceptibility. J. Hum. Genet. 56, 436–439 (2011).
Yang, J. et al. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 42, 565–569 (2010).
Morton, L.M. et al. Proposed classification of lymphoid neoplasms for epidemiologic research from the Pathology Working Group of the International Lymphoma Epidemiology Consortium (InterLymph). Blood 110, 695–708 (2007).
Turner, J.J. et al. InterLymph hierarchical classification of lymphoid neoplasms for epidemiologic research based on the WHO classification (2008): update and future directions. Blood 116, e90–e98 (2010).
Swerdlow, S., Campo, E. & Harris, N. World Health Organization Classification of Tumours of Haematopoietic and Lymphoid Tissues (IARC Press, Lyon, France, 2008).
Pritchard, J.K., Stephens, M. & Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 155, 945–959 (2000).
Price, A.L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006).
Schumacher, F.R. et al. Genome-wide association study identifies new prostate cancer susceptibility loci. Hum. Mol. Genet. 20, 3867–3875 (2011).
Siddiq, A. et al. A meta-analysis of genome-wide association studies of breast cancer identifies two novel susceptibility loci at 6q14 and 20q11. Hum. Mol. Genet. 21, 5373–5384 (2012).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Dixon, A.L. et al. A genome-wide association study of global gene expression. Nat. Genet. 39, 1202–1207 (2007).
Cheung, V.G. et al. Polymorphic cis- and trans-regulation of human gene expression. PLoS Biol. 8, e1000480 (2010).
Lee, S.H., Wray, N.R., Goddard, M.E. & Visscher, P.M. Estimating missing heritability for disease from genome-wide association studies. Am. J. Hum. Genet. 88, 294–305 (2011).
Yang, J., Lee, S.H., Goddard, M.E. & Visscher, P.M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88, 76–82 (2011).
Howlader, N. et al. SEER Cancer Statistics Review, 1975–2010. National Cancer Institute http://seer.cancer.gov/csr/1975_2010/ (2013).
Fearnhead, P. SequenceLDhot: detecting recombination hotspots. Bioinformatics 22, 3061–3066 (2006).
Fearnhead, P., Harding, R.M., Schneider, J.A., Myers, S. & Donnelly, P. Application of coalescent methods to reveal fine-scale rate variation and recombination hotspots. Genetics 167, 2067–2081 (2004).
Li, N. & Stephens, M. Modeling linkage disequilibrium and identifying recombination hotspots using single-nucleotide polymorphism data. Genetics 165, 2213–2233 (2003).
Crawford, D.C., et al. Evidence for substantial fine-scale variation in recombination rates across the human genome. Nat. Genet. 36, 700–706 (2004).
Luna, A. & Nicodemus, K.K. snp.plotter: an R-based SNP/haplotype association and linkage disequilibrium plotting package. Bioinformatics 23, 774–776 (2007).
Acknowledgements
We thank C. Allmer, E. Angelucci, A. Bigelow, S. Buehler, K. Butterbach, A. Chabrier, J.M. Conners, M. Corines, M. Cornelis, K. Corsano, H. Dykes, L. Ershler, A. Gabbas, R.P. Gallagher, R.D. Gascoyne, P. Hui, L. Irish, L. Jacobus, L. Klareskog, A.S. Lai, J. Lunde, M. McAdams, R. Montalvan, L. Padyukov, M. Rais, T. Rattle, L. Rigacci, K. Snyder, G. Specchia, M. Stagner, G. Thomas, C. Tornow, G. Wood and M. Yang. The overall GWAS project was supported by the Intramural Program of the US National Institutes of Health/National Cancer Institute. A list of support provided to individual studies appears in the Supplementary Note.
Author information
Authors and Affiliations
Contributions
J.R.C., S.I.B., S.S.W., A.N., A.R.B.-W., Q.L., G. Severi, M. Melbye, L.R.T., M.P.P., C.L., B.M.B., S.L.S., S.d.S., K.E.S., C.F.S., N.R. and S.J.C. organized and designed the study. J.R.C., L.C., L.B., A.H., P.M.B., E.A.H., S.L.S., G. Salles, C.F.S., N.R. and S.J.C. conducted and supervised the genotyping of samples. J.R.C., S.I.B., J. Vijai, Z.W., M.Y., L.C., P.I.W.d.B., D.C., J.G., D. Zhi, Y.W.A., J.H., B.M., J.S., L.L., J.-H.P., C.C.C., N.C., S.d.S., K.E.S., C.F.S., N.R. and S.J.C. contributed to the design and execution of statistical analysis. J.R.C., S.I.B., J. Vijai, H.G., J.M., S.S.W., Z.W., M.Y., L.C., A.N., D.C., A.M., C.R.F., A.J.D.R., C.L., K.E.S., C.F.S., N.R. and S.J.C. wrote the first draft of the manuscript. J.R.C., J. Vijai, H.G., J.M., S.S.W., L.C., A.N., L.B., A.M., A.R.B.-W., Q.L., G. Severi, M. Melbye, J.G., R.D.J., E.K., L.R.T., M.P.P., C.M.V., J.J.S., G.G.G., D.A., R.S.K., M.Z., K.A.B., A.Z.-J., T.M.H., B.K.L., A.J.N., A.D., Y.W.A., M.L., C.A.T., S.M.A., T.E.W., G.J.W., A.S.V., D. Zelenika, H.T., C.H., T.J.M., H.H., B.G., H.-O.A., P.M.B., J.R., M.T.S., E.A.H., W.C., P.H., L.M.M., R.K.S., L.F.T., K.E.N., N.B., Y.B., P. Boffetta, P. Brennan, L.F., M. Maynadie, A. Staines, T.L., S.C., A. Smith, E. Roman, W.R.D., K.O., A.Z., R.J.K., D.J.V., T.Z., Y.Z., T.R.H., A.K., J.T., M.C.S., J.C., J. Virtamo, S.W., E. Riboli, P.V., R.K., D.T., R.C.H.V., H.B., A.T., E.A., S.D.L., M.R., B.M.B., F.L., E.G., P.K., Y.Y., B.C.H.C., D.D.W., N.C., J.F.F., S.L.S., X.W., S.d.S., K.E.S., G. Salles, C.F.S. and N.R. conducted the epidemiological studies and contributed samples to the GWAS and/or follow-up genotyping. All authors contributed to the writing of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
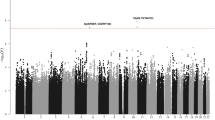
Supplementary Figure 3 Manhattan plot showing the statistical significance of the association for all genotyped SNPs in the stage 1 DLBCL GWAS.
SNPs are plotted on the x axis according to their position on each chromosome against the significance of the association on the y axis (shown as –log10 P value). The dotted line denotes P = 5 × 10–8 statistical significance.
Supplementary Figure 4 Association results, recombination hotspots and LD plots for the region 5q31.3 with DLBCL.
Top, association results of GWAS data from the stage 1 DLBCL GWAS (gray diamonds) and combined data of stages 1–3 (red diamond) are shown with –log10 (P values) (left y axis). Overlaid are the likelihood ratio statistics (right y axis) to estimate putative recombination hotspots across the region on the basis of 5 unique sets of 100 randomly selected control samples. Bottom, LD heat map based on r2 values from combined control populations for all SNPs included in the GWAS.
Supplementary Figure 5 Chromatin state dynamics of DLBCL-associated SNPs in nine human cell lines.
For details, see the Online Methods.
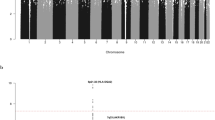
Supplementary Figure 6 Plot of estimated admixture for individuals in the NHL GWAS (stage 1).
For details, see the Online Methods. Individuals with <80% European ancestry were excluded.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7, Supplementary Tables 1–5, 7–11 and 13, and Supplementary Note. (PDF 5307 kb)
Supplementary Tables 6 and 12
Supplementary Tables 6 and 12 (XLSX 113 kb)
Rights and permissions
About this article
Cite this article
Cerhan, J., Berndt, S., Vijai, J. et al. Genome-wide association study identifies multiple susceptibility loci for diffuse large B cell lymphoma. Nat Genet 46, 1233–1238 (2014). https://doi.org/10.1038/ng.3105
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3105
This article is cited by
-
Implementation of individualised polygenic risk score analysis: a test case of a family of four
BMC Medical Genomics (2022)
-
Distinct germline genetic susceptibility profiles identified for common non-Hodgkin lymphoma subtypes
Leukemia (2022)
-
Linear and circular PVT1 in hematological malignancies and immune response: two faces of the same coin
Molecular Cancer (2020)
-
Comparative molecular cell-of-origin classification of diffuse large B-cell lymphoma based on liquid and tissue biopsies
Translational Medicine Communications (2020)
-
Genetic predisposition for multiple myeloma
Leukemia (2020)