Abstract
Osteoarthritis is the most common form of arthritis and is a major cause of pain and disability in the elderly. To search for sequence variants that confer risk of osteoarthritis of the hand, we carried out a genome-wide association study (GWAS) in subjects with severe hand osteoarthritis, using variants identified through the whole-genome sequencing of 2,230 Icelanders. We found two significantly associated loci in the Icelandic discovery set: at 15q22 (frequency of 50.7%, odds ratio (OR) = 1.51, P = 3.99 × 10−10) in the ALDH1A2 gene and at 1p31 (frequency of 0.02%, OR = 50.6, P = 9.8 × 10−10). Among the carriers of the variant at 1p31 is a family with several members in whom the risk allele segregates with osteoarthritis. The variants within the ALDH1A2 gene were confirmed in replication sets from The Netherlands and the UK, yielding an overall association of OR = 1.46 and P = 1.1 × 10−11 (rs3204689).
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
Primary accessions
Gene Expression Omnibus
Referenced accessions
Ensembl
References
Loeser, R.F., Goldring, S.R., Scanzello, C.R. & Goldring, M.B. Osteoarthritis: a disease of the joint as an organ. Arthritis Rheum. 64, 1697–1707 (2012).
Jonsson, H. et al. The inheritance of hand osteoarthritis in Iceland. Arthritis Rheum. 48, 391–395 (2003).
Spector, T.D. & MacGregor, A.J. Risk factors for osteoarthritis: genetics. Osteoarthritis Cartilage 12 (suppl. A), S39–S44 (2004).
Kraus, V.B. et al. The Genetics of Generalized Osteoarthritis (GOGO) study: study design and evaluation of osteoarthritis phenotypes. Osteoarthritis Cartilage 15, 120–127 (2007).
Haugen, I.K. et al. Prevalence, incidence and progression of hand osteoarthritis in the general population: the Framingham Osteoarthritis Study. Ann. Rheum. Dis. 70, 1581–1586 (2011).
Kerkhof, H.J. et al. A genome-wide association study identifies an osteoarthritis susceptibility locus on chromosome 7q22. Arthritis Rheum. 62, 499–510 (2010).
Evangelou, E. et al. Meta-analysis of genome-wide association studies confirms a susceptibility locus for knee osteoarthritis on chromosome 7q22. Ann. Rheum. Dis. 70, 349–355 (2011).
Day-Williams, A.G. et al. A variant in MCF2L is associated with osteoarthritis. Am. J. Hum. Genet. 89, 446–450 (2011).
Castaño Betancourt, M.C. et al. Genome-wide association and functional studies identify the DOT1L gene to be involved in cartilage thickness and hip osteoarthritis. Proc. Natl. Acad. Sci. USA 109, 8218–8223 (2012).
arcOGEN Consortium. Identification of new susceptibility loci for osteoarthritis (arcOGEN): a genome-wide association study. Lancet 380, 815–823 (2012).
Miyamoto, Y. et al. Common variants in DVWA on chromosome 3p24.3 are associated with susceptibility to knee osteoarthritis. Nat. Genet. 40, 994–998 (2008).
Nakajima, M. et al. New sequence variants in HLA class II/III region associated with susceptibility to knee osteoarthritis identified by genome-wide association study. PLoS ONE 5, e9723 (2010).
Evangelou, E. et al. A meta-analysis of genome-wide association studies identifies novel variants associated with osteoarthritis of the hip. Ann. Rheum. Dis. 10.1136/annrheumdis-2012-203114 (4 September 2013).
Riyazi, N. et al. Evidence for familial aggregation of hand, hip, and spine but not knee osteoarthritis in siblings with multiple joint involvement: the GARP study. Ann. Rheum. Dis. 64, 438–443 (2005).
Schoenmaker, M. et al. Evidence of genetic enrichment for exceptional survival using a family approach: the Leiden Longevity Study. Eur. J. Hum. Genet. 14, 79–84 (2006).
Hofman, A. et al. The Rotterdam Study: 2012 objectives and design update. Eur. J. Epidemiol. 26, 657–686 (2011).
Spector, T.D. & Williams, F.M. The UK Adult Twin Registry (TwinsUK). Twin Res. Hum. Genet. 9, 899–906 (2006).
Hart, D.J. & Spector, T.D. Cigarette smoking and risk of osteoarthritis in women in the general population: the Chingford study. Ann. Rheum. Dis. 52, 93–96 (1993).
Dahaghin, S. et al. Does hand osteoarthritis predict future hip or knee osteoarthritis? Arthritis Rheum. 52, 3520–3527 (2005).
Jonsson, H. et al. Hand osteoarthritis severity is associated with total knee joint replacements independently of BMI. The Ages-Reykjavik Study. Open Rheumatol. J. 5, 7–12 (2011).
Valdes, A.M. et al. Involvement of different risk factors in clinically severe large joint osteoarthritis according to the presence of hand interphalangeal nodes. Arthritis Rheum. 62, 2688–2695 (2010).
Rhinn, M. & Dolle, P. Retinoic acid signalling during development. Development 139, 843–858 (2012).
Gudas, L.J. Emerging roles for retinoids in regeneration and differentiation in normal and disease states. Biochim. Biophys. Acta 1821, 213–221 (2012).
Underhill, T.M. & Weston, A.D. Retinoids and their receptors in skeletal development. Microsc. Res. Tech. 43, 137–155 (1998).
Weston, A.D., Hoffman, L.M. & Underhill, T.M. Revisiting the role of retinoid signaling in skeletal development. Birth Defects Res. C Embryo Today 69, 156–173 (2003).
Laue, K. et al. Craniosynostosis and multiple skeletal anomalies in humans and zebrafish result from a defect in the localized degradation of retinoic acid. Am. J. Hum. Genet. 89, 595–606 (2011).
Theodosiou, M., Laudet, V. & Schubert, M. From carrot to clinic: an overview of the retinoic acid signaling pathway. Cell. Mol. Life Sci. 67, 1423–1445 (2010).
Al Tanoury, Z., Piskunov, A. & Rochette-Egly, C. Vitamin A and retinoid signaling: genomic and non-genomic effects. J. Lipid Res. 54, 1761–1775 (2013).
Pittlik, S. & Begemann, G. New sources of retinoic acid synthesis revealed by live imaging of an Aldh1a2-GFP reporter fusion protein throughout zebrafish development. Dev. Dyn. 241, 1205–1216 (2012).
Duester, G. Retinoic acid synthesis and signaling during early organogenesis. Cell 134, 921–931 (2008).
Niederreither, K., Subbarayan, V., Dolle, P. & Chambon, P. Embryonic retinoic acid synthesis is essential for early mouse post-implantation development. Nat. Genet. 21, 444–448 (1999).
Zhao, X. et al. Retinoic acid promotes limb induction through effects on body axis extension but is unnecessary for limb patterning. Curr. Biol. 19, 1050–1057 (2009).
Taylor, J. et al. ESPERR: learning strong and weak signals in genomic sequence alignments to identify functional elements. Genome Res. 16, 1596–1604 (2006).
ENCODE Project Consortium. A users guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol. 9, e1001046 (2011).
Kong, A. et al. Fine-scale recombination rate differences between sexes, populations and individuals. Nature 467, 1099–1103 (2010).
Pruim, R.J. et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 26, 2336–2337 (2010).
Mantel, N. & Haenszel, W. Statistical aspects of the analysis of data from retrospective studies of disease. J. Natl. Cancer Inst. 22, 719–748 (1959).
Styrkarsdottir, U. et al. Nonsense mutation in the LGR4 gene is associated with several human diseases and other traits. Nature 497, 517–520 (2013).
Kong, A. et al. Detection of sharing by descent, long-range phasing and haplotype imputation. Nat. Genet. 40, 1068–1075 (2008).
Kong, A. et al. Parental origin of sequence variants associated with complex diseases. Nature 462, 868–874 (2009).
Devlin, B. & Roeder, K. Genomic control for association studies. Biometrics 55, 997–1004 (1999).
Kerkhof, H.J.M. et al. Recommendations for standardization and phenotype definitions in genetic studies of osteoarthritis: the TREAT-OA consortium. Osteoarthritis Cartilage 19, 254–264 (2011).
Panoutsopoulou, K. et al. Insights into the genetic architecture of osteoarthritis from stage 1 of the arcOGEN study. Ann. Rheum. Dis. 70, 864–867 (2011).
Raine, E.V.A., Dodd, A., Reynard, L. & Loughlin, J. Allelic expression analysis of the osteoarthritis susceptibility gene COL11A1 in human joint tissues. BMC Musculoskelet. Disord. 14, 85 (2013).
Bos, S.D. et al. Increased type II deiodinase protein in OA-affected cartilage and allelic imbalance of OA risk polymorphism rs225014 at DIO2 in human OA joint tissues. Ann. Rheum. Dis. 71, 1254–1258 (2012).
Emilsson, V. et al. Genetics of gene expression and its effect on disease. Nature 452, 423–428 (2008).
Stefansson, H. et al. A common inversion under selection in Europeans. Nat. Genet. 37, 129–137 (2005).
Meyer, L.R. et al. The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res. 41, D64–D69 (2013).
Rosenbloom, K.R. et al. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res. 41, D56–D63 (2013).
Acknowledgements
We thank the subjects of the deCODE study, the Rotterdam study, the GARP, LLS, RAAK and PAPRIKA studies, the TwinsUK and Chingford studies, and the MDC study for their valuable participation. This work was supported by European Union Framework Programme 7 grant 200800 TREAT-OA. Full acknowledgments are given in the Supplementary Note.
Author information
Authors and Affiliations
Consortia
Contributions
The study was designed and results were interpreted by U.S., G.T., I.M., U.T., I.J., H.J. and K.S. Phenotype data and Icelandic subject recruitment were coordinated and managed by T.I. and H.J. D.H., S.M., N.K.A., T.S. and A.M.V. coordinated, managed, genotyped and analyzed the TwinsUK and Chingford cohort samples. M.B., M.K., P.E.S. and I.M. coordinated, managed, genotyped and analyzed the samples from the GARP, LLS, PAPRIKA and RAAK studies. A.H., F.R., A.U., S.B.-Z. and J.v.M. coordinated, managed, genotyped and analyzed the samples from the Rotterdam cohorts. S.L. coordinated and managed the samples from the MDC study. E.E. undertook the meta-analysis for the TreatOA consortium. G.M., U.T., G.T. and U.S. coordinated sequence data analysis, imputation and association analysis in the Icelandic data set. O.T.M. and S.A.G. performed exome sequencing and analysis. S.A.G., A.J. and A.S. carried out bioinformatics analysis for 1p31. Shared haplotype analysis was performed by M.L.F. and A.K. Sanger sequencing of Icelandic samples and Centaurus genotyping of Icelandic and Swedish samples were carried out and analyzed by H.T.H. and U.S. Expression experiments were carried out and analyzed by N.B., A.M.S., R.G.H.H.N., A.J., A.S., F.F.E. and M.T. The manuscript was drafted by U.S., G.T. and K.S. All authors contributed to the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
U.S., G.T., H.T.H., A.J., A.S., S.A.G., M.L.F., A.K., O.T.M., G.M., U.T., I.J. and K.S. are employed by deCODE Genetics/Amgen.
Additional information
Full list of members and affiliations appears in the Supplementary Note.
Full list of members and affiliations appears in the Supplementary Note.
Integrated supplementary information
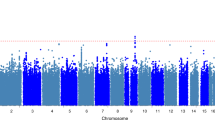
Supplementary Figure 1 Manhattan plot of the discovery GWAS.
P values (–log10) are plotted against their respective positions on each chromosome. P = 5 × 10−8 is indicated by the horizontal pink line.
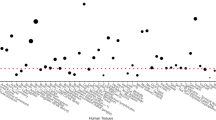
Supplementary Figure 2 Differential allelic expression of ALDH1A2 transcript in cartilage according to genotype at rs3204689.
Allelic imbalance depicted by allele ratios (G/C) plotted as log2 values for 22 individuals heterozygous for the rs3204689 SNP. Cartilage was isolated from the hip or knee joints of individuals with osteoarthritis. (a) Preserved (macroscopically normal) articular cartilage samples (N = 15). (b) Osteoarthritis-affected articular cartilage samples (N = 17). *P < 0.0005, **P < 0.0001, ***P < 0.00001.
Supplementary Figure 3 Genes in the 1p31 locus region.
Locations of genes in the region on 1p31 that is shared by all the affected carriers and the locations of the three associated markers and the LD blocks are shown. The genes are LINC00466 (long intergenic non-protein-coding RNA 466), FOXD3 (forkhead box D3), ALG6 (α-1,3-glucosyltransferase), ITGB3BP (integrin β3–binding protein), EFCAB7 (EF-hand calcium-binding domain 7), PGM1 (phosphoglucomutase 1) and ROR1 (receptor tyrosine kinase–like orphan receptor 1), and the pseudogene is DLEU2L (deleted in lymphocytic leukemia 2-like). All positions are in NCBI Build 36 coordinates. The plot was created using a stand-alone version of LocusZoom software35 (http://csg.sph.umich.edu/locuszoom/).
Supplementary information
Supplementary Text and Figures
Supplementary Note, Supplementary Tables 1–6 and Supplementary Figures 1–3 (PDF 1727 kb)
Rights and permissions
About this article
Cite this article
Styrkarsdottir, U., Thorleifsson, G., Helgadottir, H. et al. Severe osteoarthritis of the hand associates with common variants within the ALDH1A2 gene and with rare variants at 1p31. Nat Genet 46, 498–502 (2014). https://doi.org/10.1038/ng.2957
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2957
This article is cited by
-
Erosive Hand Osteoarthritis: Recent Advances and Future Treatments
Current Rheumatology Reports (2024)
-
Identification of candidate enhancers controlling the transcriptome during the formation of interphalangeal joints
Scientific Reports (2022)
-
An eQTL variant of ALDH1A2 is associated with Kashin-Beck disease in Chinese population
Journal of Bone and Mineral Metabolism (2022)
-
The Genetic Epidemiology of Joint Shape and the Development of Osteoarthritis
Calcified Tissue International (2021)
-
Current Epidemiology and Risk Factors for the Development of Hand Osteoarthritis
Current Rheumatology Reports (2021)