Abstract
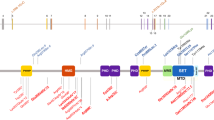
Overgrowth disorders are a heterogeneous group of conditions characterized by increased growth parameters and other variable clinical features such as intellectual disability and facial dysmorphism1. To identify new causes of human overgrowth, we performed exome sequencing in ten proband-parent trios and detected two de novo DNMT3A mutations. We identified 11 additional de novo mutations by sequencing DNMT3A in a further 142 individuals with overgrowth. The mutations alter residues in functional DNMT3A domains, and protein modeling suggests that they interfere with domain-domain interactions and histone binding. Similar mutations were not present in 1,000 UK population controls (13/152 cases versus 0/1,000 controls; P < 0.0001). Mutation carriers had a distinctive facial appearance, intellectual disability and greater height. DNMT3A encodes a DNA methyltransferase essential for establishing methylation during embryogenesis and is commonly somatically mutated in acute myeloid leukemia2,3,4. Thus, DNMT3A joins an emerging group of epigenetic DNA- and histone-modifying genes associated with both developmental growth disorders and hematological malignancies5.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Change history
25 April 2014
In the version of this article initially published, the protein alterations for three cases (COG1770, COG1670 and COG0141) were listed incorrectly in Table 1. The correct protein alterations for these three cases are p.Ile310Asn, p.Ser312fs and p.Gly532Ser, respectively. These errors have been corrected in the HTML and PDF versions of the article.
References
Tatton-Brown, K. & Weksberg, R. Molecular mechanisms of childhood overgrowth. Am. J. Med. Genet. C. Semin. Med. Genet. 163C, 71–75 (2013).
Jurkowska, R.Z., Jurkowski, T.P. & Jeltsch, A. Structure and function of mammalian DNA methyltransferases. ChemBioChem 12, 206–222 (2011).
Ley, T.J. et al. DNMT3A mutations in acute myeloid leukemia. N. Engl. J. Med. 363, 2424–2433 (2010).
Yan, X.J. et al. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat. Genet. 43, 309–315 (2011).
Tatton-Brown, K. & Rahman, N. The NSD1 and EZH2 overgrowth genes, similarities and differences. Am. J. Med. Genet. C. Semin. Med. Genet. 163C, 86–91 (2013).
Durand, C. & Rappold, G.A. Height matters—from monogenic disorders to normal variation. Nat. Rev. Endocrinol. 9, 171–177 (2013).
Rush, M. et al. Targeting of EZH2 to a defined genomic site is sufficient for recruitment of Dnmt3a but not de novo DNA methylation. Epigenetics 4, 404–414 (2009).
Tatton-Brown, K. et al. Germline mutations in the oncogene EZH2 cause Weaver syndrome and increased human height. Oncotarget 2, 1127–1133 (2011).
Freeman, J.V et al. Cross sectional stature and weight reference curves for the UK, 1990. Arch. Dis. Child. 73, 17–24 (1995).
Okano, M., Bell, D.W., Haber, D.A. & Li, E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247–257 (1999).
Smallwood, S.A. & Kelsey, G. De novo DNA methylation: a germ cell perspective. Trends Genet. 28, 33–42 (2012).
Kaneda, M. et al. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature 429, 900–903 (2004).
Chedin, F., Lieber, M.R. & Hsieh, C.L. The DNA methyltransferase–like protein DNMT3L stimulates de novo methylation by Dnmt3a. Proc. Natl. Acad. Sci. USA 99, 16916–16921 (2002).
Hata, K., Okano, M., Lei, H. & Li, E. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development 129, 1983–1993 (2002).
Suetake, I., Shinozaki, F., Miyagawa, J., Takeshima, H. & Tajima, S. DNMT3L stimulates the DNA methylation activity of Dnmt3a and Dnmt3b through a direct interaction. J. Biol. Chem. 279, 27816–27823 (2004).
Abdel-Wahab, O. & Levine, R.L. Mutations in epigenetic modifiers in the pathogenesis and therapy of acute myeloid leukemia. Blood 121, 3563–3572 (2013).
Marcucci, G. et al. Age-related prognostic impact of different types of DNMT3A mutations in adults with primary cytogenetically normal acute myeloid leukemia. J. Clin. Oncol. 30, 742–750 (2012).
Nikoloski, G., van der Reijden, B.A. & Jansen, J.H. Mutations in epigenetic regulators in myelodysplastic syndromes. Int. J. Hematol. 95, 8–16 (2012).
Kim, S.J. et al. A DNMT3A mutation common in AML exhibits dominant-negative effects in murine ES cells. Blood 122, 4086–4089 (2013).
Otani, J. et al. Structural basis for recognition of H3K4 methylation status by the DNA methyltransferase 3A ATRX-DNMT3-DNMT3L domain. EMBO Rep. 10, 1235–1241 (2009).
Tatton-Brown, K. et al. Genotype-phenotype associations in Sotos syndrome: an analysis of 266 individuals with NSD1 aberrations. Am. J. Hum. Genet. 77, 193–204 (2005).
Lund, K., Adams, P.D. & Copland, M. EZH2 in normal and malignant hematopoiesis. Leukemia 28, 44–49 (2014).
Cerveira, N. et al. Frequency of NUP98-NSD1 fusion transcript in childhood acute myeloid leukaemia. Leukemia 17, 2244–2247 (2003).
Cao, R. & Zhang, Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr. Opin. Genet. Dev. 14, 155–164 (2004).
Morishita, M. & di Luccio, E. Structural insights into the regulation and the recognition of histone marks by the SET domain of NSD1. Biochem. Biophys. Res. Commun. 412, 214–219 (2011).
Hoischen, A. et al. De novo nonsense mutations in ASXL1 cause Bohring-Opitz syndrome. Nat. Genet. 43, 729–731 (2011).
Lunter, G. & Goodson, M. Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res. 21, 936–939 (2011).
Rimmer, A., Mathieson, I., Lunter, G. & McVean, G. Platypus: an integrated variant caller, http://www.well.ox.ac.uk/platypus(2012).
Acknowledgements
We thank the families for their participation in our research and the physicians and nurses who recruited them. Samples were collected through the Childhood Overgrowth Collaboration; a full list of collaborators is presented in the Supplementary Note. We are grateful to M. Warren-Perry, D. Dudakia and J. Bull for assistance in recruitment and to E. Moran (New York University Hospital for Joint Diseases) and A. Murray (University Hospital of Wales) for their clinical input for COG1770 and COG0109, respectively. We thank A. Strydom for assistance in preparing the manuscript. We are grateful to G. Lunter and M. Münz (Wellcome Trust Centre for Human Genetics, Oxford University) for their contributions to the development of the custom annotation tool SAVANT. We acknowledge use of services provided by the Institute of Cancer Research Genetics Core Facility, which is managed by S.H. and N.R. We acknowledge National Health Service (NHS) funding to the Royal Marsden/Institute of Cancer Research National Institute for Health Research (NIHR) Biomedical Research Centre. We also thank Mariani Foundation Milan for supporting the clinical activity of Genetica Clinica Pediatrica, Fondazione MBBM, AO San Gerardo Monza. This research was supported by the Wellcome Trust (100210/Z/12/Z) and by the Institute of Cancer Research, London.
Author information
Authors and Affiliations
Consortia
Contributions
S.S., E. Ramsay., S.d.V.D., S.H. and E.O. undertook the molecular analyses. E. Ruark undertook the bioinformatics analyses. A.Z. coordinated recruitment. L.A., D.B., T.D., B.G., D.G., T.H., A.K., D.T.P., A.S., I.K.T., L.V.M., N.Y. and K.T.-B. collected samples and undertook phenotyping. J.H. and R.v.M. undertook the protein modeling. N.R. and K.T.-B. designed and oversaw the project and wrote the manuscript with input from other authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
A full list of members appears in the Supplementary Note.
Integrated supplementary information
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1 and 2, Supplementary Figure 1 and Supplementary Note (PDF 429 kb)
Rights and permissions
About this article
Cite this article
Tatton-Brown, K., Seal, S., Ruark, E. et al. Mutations in the DNA methyltransferase gene DNMT3A cause an overgrowth syndrome with intellectual disability. Nat Genet 46, 385–388 (2014). https://doi.org/10.1038/ng.2917
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2917
This article is cited by
-
O-GlcNAcylation: the sweet side of epigenetics
Epigenetics & Chromatin (2023)
-
Methylation signatures in clinically variable syndromic disorders: a familial DNMT3A variant in two adults with Tatton-Brown–Rahman syndrome
European Journal of Human Genetics (2023)
-
Regulation, functions and transmission of bivalent chromatin during mammalian development
Nature Reviews Molecular Cell Biology (2023)
-
Differential expression of m5C RNA methyltransferase genes NSUN6 and NSUN7 in Alzheimer’s disease and traumatic brain injury
Molecular Neurobiology (2023)
-
Histone H3K36me2 and H3K36me3 form a chromatin platform essential for DNMT3A-dependent DNA methylation in mouse oocytes
Nature Communications (2022)