Abstract
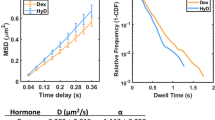
Transcription factors mediate gene regulation by site-specific binding to chromosomal operators. It is commonly assumed that the level of repression is determined solely by the equilibrium binding of a repressor to its operator. However, this assumption has not been possible to test in living cells. Here we have developed a single-molecule chase assay to measure how long an individual transcription factor molecule remains bound at a specific chromosomal operator site. We find that the lac repressor dimer stays bound on average 5 min at the native lac operator in Escherichia coli and that a stronger operator results in a slower dissociation rate but a similar association rate. Our findings do not support the simple equilibrium model. The discrepancy with this model can, for example, be accounted for by considering that transcription initiation drives the system out of equilibrium. Such effects need to be considered when predicting gene activity from transcription factor binding strengths.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Bintu, L. et al. Transcriptional regulation by the numbers: applications. Curr. Opin. Genet. Dev. 15, 125–135 (2005).
Oehler, S., Amouyal, M., Kolkhof, P., von Wilcken-Bergmann, B. & Muller-Hill, B. Quality and position of the three lac operators of E. coli define efficiency of repression. EMBO J. 13, 3348–3355 (1994).
Oehler, S., Eismann, E.R., Kramer, H. & Muller-Hill, B. The three operators of the lac operon cooperate in repression. EMBO J. 9, 973–979 (1990).
Choi, P.J., Cai, L., Frieda, K. & Xie, X.S. A stochastic single-molecule event triggers phenotype switching of a bacterial cell. Science 322, 442–446 (2008).
Hammar, P. et al. The Lac repressor displays facilitated diffusion in living cells. Science 336, 1595–1598 (2012).
Riggs, A.D., Bourgeois, S. & Cohn, M. The lac repressor-operator interaction. 3. Kinetic studies. J. Mol. Biol. 53, 401–417 (1970).
Chang, W.I., Barrera, P. & Matthews, K.S. Identification and characterization of aspartate residues that play key roles in the allosteric regulation of a transcription factor: aspartate 274 is essential for inducer binding in lac repressor. Biochemistry 33, 3607–3616 (1994).
Bell, C.E. & Lewis, M. A closer view of the conformation of the Lac repressor bound to operator. Nat. Struct. Biol. 7, 209–214 (2000).
Bell, C.E. & Lewis, M. Crystallographic analysis of Lac repressor bound to natural operator O1. J. Mol. Biol. 312, 921–926 (2001).
Garcia, H.G. et al. Operator sequence alters gene expression independently of transcription factor occupancy in bacteria. Cell Reports 2, 150–161 (2012).
Sanchez, A., Osborne, M.L., Friedman, L.J., Kondev, J. & Gelles, J. Mechanism of transcriptional repression at a bacterial promoter by analysis of single molecules. EMBO J. 30, 3940–3946 (2011).
Kuhlman, T.E. & Cox, E.C. Gene location and DNA density determine transcription factor distributions in Escherichia coli. Mol. Syst. Biol. 8, 610 (2012).
Hermsen, R., ten Wolde, P.R. & Teichmann, S. Chance and necessity in chromosomal gene distributions. Trends Genet. 24, 216–219 (2008).
Vignali, M., Hassan, A.H., Neely, K.E. & Workman, J.L. ATP-dependent chromatin-remodeling complexes. Mol. Cell. Biol. 20, 1899–1910 (2000).
Seila, A.C., Core, L.J., Lis, J.T. & Sharp, P.A. Divergent transcription: a new feature of active promoters. Cell Cycle 8, 2557–2564 (2009).
Datsenko, K.A. & Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97, 6640–6645 (2000).
Link, A.J., Phillips, D. & Church, G.M. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 179, 6228–6237 (1997).
Ullman, G. et al. High-throughput gene expression analysis at the level of single proteins using a microfluidic turbidostat and automated cell tracking. Phil. Trans. R. Soc. Lond. B 368, 20120025 (2013).
Edelstein, A., Amodaj, N., Hoover, K., Vale, R. & Stuurman, N. Computer control of microscopes using microManager. Curr. Protoc. Mol. Biol. Chapter 14, Unit14.20 (2010).
Olivo-Marin, J.C. Extraction of spots in biological images using multiscale products. Pattern Recognit. 35, 1989–1996 (2002).
Sadler, B.M. & Swami, A. Analysis of multiscale products for step detection and estimation. IEEE Trans. Inf. Theory 45, 1043–1051 (1999).
Nagai, T. et al. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 20, 87–90 (2002).
Elf, J., Li, G.W. & Xie, X.S. Probing transcription factor dynamics at the single-molecule level in a living cell. Science 316, 1191–1194 (2007).
Taniguchi, Y. et al. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science 329, 533–538 (2010).
Yu, J., Xiao, J., Ren, X., Lao, K. & Xie, X.S. Probing gene expression in live cells, one protein molecule at a time. Science 311, 1600–1603 (2006).
Acknowledgements
We thank K.S. Matthews for advice on LacI mutants, G.-W. Li for helpful comments, X.S. Xie (Harvard University) for the MalI-Venus strain and I. Barkefors for critical reading of the manuscript. This work was supported by the European Research Council (ERC), the Knut and Alice Wallenberg Foundation (KAW), Vetenskapsrådet (VR), the Foundation for Strategic Research (SSF) and the Göran Gustafsson Foundation.
Author information
Authors and Affiliations
Contributions
J.E. conceived the project, J.E. and P.H. conceived the chase method, M.W. designed the microfluidics chip, P.H. and P.L. generated strains, P.H., M.W., Ö.B., F.P. and P.L. performed experiments, F.P., G.U., D.F. and M.W. developed and implemented analysis routines, P.H., F.P., M.W., G.U., D.F. and Ö.B. analyzed the data, J.E. and D.F. developed theoretical models, and J.E., P.H., D.F., M.W. and F.P. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Single-molecule chase assay for dissociation.
(a) Western blot of strain JE127 harboring pBAD24-lacI. While plasmid-expressed LacI was immediately upregulated by the addition of arabinose, chromosomal LacIs-Venus remained unaffected. Using ImageJ (supplementary reference 2) the excess of competitor (LacI over LacIs-Venus) was quantified for each time point to be 4× (at 0 min), 5× (1 min), 12× (3 min) and >20× (5 and 10 min). (b) Raw data of the different dissociation rate measurements shown in Figure 2a and Supplementary Figure 4b—each data set is fitted including an initial association of fluorescent molecules as described in the Online Methods. For each strain/condition all series are fitted to a fixed generation time (i.e., time between divisions, which is 26 min at 37 °C (Supplementary Fig. 3a) and 70 min at 25 °C (data not shown)), a common toff and individual parameters for start and end levels. (c) LacIs-Venus binding to lacOsym at 37 °C; in black circles (as in Fig. 2a)—pBAD24-lacI, switch from IPTG to arabinose; in green squares—pBAD24-lacI, switch from IPTG to IPTG + arabinose; in red triangles—pBAD24-xylR, switch from IPTG to IPTG + arabinose.
Supplementary Figure 2 Fast switching kinetics as demonstrated by IPTG response.
(a) Images show binding of LacI-Venus to lacO1 (at 37 °C), with automatically detected localized spots encircled, before (0) and 0.4, 2.8 and 5.2 s after removal of IPTG. Scale bar = 4 μm. As described in the Supplementary Note, a 2-s delay was subtracted from the time points to account for the time delay from the medium switch. (b) The corresponding dissociation curves from lacO1 and lacOsym operators. (c) Association rates to lacOsym measured with increasing competing concentration of the anti-inducer ONPF. (d) Steady-state binding (number of spots per cell after 15-min incubation with IPTG at different concentrations) to lacOsym.
Supplementary Figure 3 Average number of operator sites.
The number of lac operator sites per cell increases at replication, decreases at division and can be detected as specifically bound LacI-Venus molecules. (a) The observed frequency (number of molecules cell-1 min-1 μm-1) of specifically bound LacI-Venus molecules mapped onto the major axis (horizontal) of the cells over time (vertical, from top to bottom). The exponentially expanding black lines indicate the average apex position of the poles of the cells, and the red horizontal lines indicate the average division times. Cells are synchronized at the time they achieve a reference length (4.25 μm). (b) The contribution of spots detected in a strain in which all operator sites have been removed (same scale as in a). (c) Same as (a) but with cells synchronized according to time of division.
Supplementary Figure 4 Roadblock effect for association to lacOsym and lacO1 and dissociation from lacOsym.
(a) Association to lacOsym with and without roadblock on one side of the operator; the roadblock effect is f = 1.35 ± 0.04. (b) Dissociation with and without roadblock; the same data as in Supplementary Figure 1b (bottom right panel) and fitted as described in the Supplementary Note; the roadblock effect is f = 1.25 ± 0.09. (c) Association in a strain with two lacO1 operators with binding sites for two TetR roadblocks in between. Association curves with and without TetR expressed are shown. The roadblock effect is f = 1.16 ± 0.03. (a–c) We verified that the TetR roadblock binds >99% of the time by comparing β-galactosidase activity ± aTC in a strain (JE125; Supplementary Fig. 7b) where lacO1 is replaced by the tet operator tetO2 and TetR is chromosomally and highly expressed as in the kinetic experiments. The repression ratio is 220 ± 26 (mean ± s.d.). As a control the experiment was repeated without TetR expressed (strain JE112), and here the repression ratio is 1.0 ± 0.35 (mean ± s.d.). Comment: the rates for binding to two operator sites in (c) are not a factor of two higher as compared to Figure 2b as under these experimental conditions, i.e., exponential growth at 37 °C in the microfluidic device, the fraction of mature Venus is on the order of 0.25 and many sites are occupied by non-fluorescent LacI-Venus.
Supplementary Figure 5 Spatial distribution of non-specifically bound LacI-Venus molecules in cells between 4.6 μm and 5.1 μm.
(a) The distribution of LacI-Venus molecules observed at 2-ms exposures in strains without specific operator sites. The top panel shows the distribution of LacI42-Venus (n = 20,863 cells with 〈s〉 = 0.26 detected spots per cell), in which the DNA binding domain of LacI has been deleted. The following panels (from top to bottom) show strains expressing LacI-Venus from loci proximate to the origin of replication (n = 15,538, 〈s〉 = 0.77), mid-replicore (n = 13,572, 〈s〉 = 0.65) and the terminus (n = 19,023, 〈s〉 = 0.63), respectively. (b) Localization of chromosomal gene loci as determined by the specific binding to operator sites. Top, origin seen as the binding of plasmid-expressed LacI-Venus to an array of lacO1 operators (n = 5,453, 〈s〉 = 1.55). Middle, mid-replicore seen as the binding of chromosomal LacI-Venus to single lacO1 (n = 10,942, 〈s〉 = 0.5) (data the same as in Supplementary Fig. 3a). Bottom, terminus seen as the binding of chromosomal MalI-Venus to two colocalized malO sites (n = 12,648, 〈s〉 = 0.8. (c) Distribution of LacI42-Venus without (top) (n = 517, 〈s〉 = 1.35) and LacI-Venus with (bottom) (n = 1,054, 〈s〉 = 1.7), DNA binding domain; expressed from the mid-replicore region and imaged 40 min after fixation with formaldehyde. (d) Same as (c), although with data pooled from the whole data set and thus a broader range of cell lengths (2.3–5.6 μm) (top) (n = 17,579, 〈s〉 = 0.96) (bottom) (n = 15,175, 〈s〉 = 1.38). (e) Time lapse showing how non-specifically binding LacI-Venus molecules are immobilized by formaldehyde treatment; the detection of near-diffraction limited spots increases with time when imaging is done using long exposure times (blue squares), and the rate of diffusion decreases on the same timescale as measured by fast tracking (3.5 ms between frames). (f) The growth of single cells as a function of time (25 s between frames) before and after (vertical red dashed line) formaldehyde treatment. The data show the average of 230 individual cell traces. The vertical black continuous line marks the point of growth arrest as found by regression analysis. Inset, a subset of the cell traces (from a single trap in the microfluidic chip) and the fitting to each one of them is shown with the same axis labels as in the main figure.
Supplementary Figure 6 Tracking of LacI-Venus in cells between 2 and 3 μm.
(a) Trajectories longer than 30 ms at a sampling rate of 330 Hz with 1-ms exposures in a strain without specific operator sites, mapped onto a unit cell (425 trajectories from approximately 500 cells). Five trajectories were randomly chosen and highlighted. (b) The distribution of detected spots mapped onto a unit cell (117,891 points from approximately 500 cells). (c) MSD analysis for the two-dimensional trajectory data presented in (a). The analysis was performed prior to mapping coordinates onto the unit cell. The offset indicates a localization accuracy of 40 nm. (d) A histogram showing the distribution for step lengths in two dimensions for the diffusing molecules during a time of 9 ms, i.e,. for trajectories subsampled by a factor of 3.
Supplementary Figure 7 Strains used in this study.
Those marked with an asterisk were previously published (supplementary reference 1). (a) Strains used in microscopy experiments and (b) strains used in β-galactosidase experiments.
Supplementary Figure 8 Calibration of LacI-Venus expression levels.
(a) From left to right: total fluorescence in strains (Supplementary Fig. 7) having LacI-Venus being autorepressed at the sequence lacO1 (JE116) and the sequence lacOsym (JE101) then in two strains having LacI-Venus and no autorepression (JE120, JE124) and a strain lacking Venus (BW25993). Scale bar = 4 μm. (b) Western blot using antibody against LacI in the corresponding strains. From left to right: JE116, JE101, JE120, JE124 and BW25993.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8, Supplementary Table 1 and Supplementary Note (PDF 4833 kb)
Rights and permissions
About this article
Cite this article
Hammar, P., Walldén, M., Fange, D. et al. Direct measurement of transcription factor dissociation excludes a simple operator occupancy model for gene regulation. Nat Genet 46, 405–408 (2014). https://doi.org/10.1038/ng.2905
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2905
This article is cited by
-
Gene expression model inference from snapshot RNA data using Bayesian non-parametrics
Nature Computational Science (2023)
-
Imaging-based screens of pool-synthesized cell libraries
Nature Methods (2021)
-
Time-resolved imaging-based CRISPRi screening
Nature Methods (2020)
-
Inferring quantity and qualities of superimposed reaction rates from single molecule survival time distributions
Scientific Reports (2020)
-
Design and Application of an Artificial Hybrid PromoterPluxI-lacOin Genetic Circuit to Achieve Lower Basal Expression Level
Applied Biochemistry and Biotechnology (2020)