Abstract
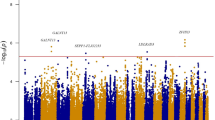
Genome-wide association studies have shown that variation in MTNR1B (melatonin receptor 1B) is associated with insulin and glucose concentrations. Here we show that the risk genotype of this SNP predicts future type 2 diabetes (T2D) in two large prospective studies. Specifically, the risk genotype was associated with impairment of early insulin response to both oral and intravenous glucose and with faster deterioration of insulin secretion over time. We also show that the MTNR1B mRNA is expressed in human islets, and immunocytochemistry confirms that it is primarily localized in β cells in islets. Nondiabetic individuals carrying the risk allele and individuals with T2D showed increased expression of the receptor in islets. Insulin release from clonal β cells in response to glucose was inhibited in the presence of melatonin. These data suggest that the circulating hormone melatonin, which is predominantly released from the pineal gland in the brain, is involved in the pathogenesis of T2D. Given the increased expression of MTNR1B in individuals at risk of T2D, the pathogenic effects are likely exerted via a direct inhibitory effect on β cells. In view of these results, blocking the melatonin ligand-receptor system could be a therapeutic avenue in T2D.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Diabetes Genetics Initiative of Broad Institute of Harvard and MIT, Lund University, & Novartis Institutes of BioMedical Research, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 316, 1331–1336 (2007).
Scott, L.J. et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 316, 1341–1345 (2007).
Sladek, R. et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 445, 881–885 (2007).
Zeggini, E. et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat. Genet. 40, 638–645 (2008).
Zeggini, E. et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science 316, 1336–1341 (2007).
Prokopenko, I. et al. Variants in MTNR1B influence fasting glucose levels and risk of type 2 diabetes. Nat. Genet. advance online publication, doi:10.1038/ng.290 (7 December 2008).
Peschke, E. Melatonin, endocrine pancreas and diabetes. J. Pineal Res. 44, 26–40 (2008).
Kvetnoy, I.M. Extrapineal melatonin: location and role within diffuse neuroendocrine system. Histochem. J. 31, 1–12 (1999).
Boden, G., Ruiz, J., Urbain, J.L. & Chen, X. Evidence for a circadian rhythm of insulin secretion. Am. J. Physiol. 271, E246–E252 (1996).
Pandi-Perumal, S.R. et al. Physiological effects of melatonin: role of melatonin receptors and signal transduction pathways. Prog. Neurobiol. 85, 335–353 (2008).
Muhlbauer, E. & Peschke, E. Evidence for the expression of both the MT1- and in addition, the MT2-melatonin receptor, in the rat pancreas, islet and beta-cell. J. Pineal Res. 42, 105–106 (2007).
Ramracheya, R.D. et al. Function and expression of melatonin receptors on human pancreatic islets. J. Pineal Res. 44, 273–279 (2008).
Valle, T. et al. Mapping genes for NIDDM. Design of the Finland-United States Investigation of NIDDM Genetics (FUSION) Study. Diabetes Care 21, 949–958 (1998).
Eriksson, J.G., Osmond, C., Kajantie, E., Forsen, T.J. & Barker, D.J. Patterns of growth among children who later develop type 2 diabetes or its risk factors. Diabetologia 49, 2853–2858 (2006).
Marselli, L. et al. Gene expression of purified beta-cell tissue obtained from human pancreas with laser capture microdissection. J. Clin. Endocrinol. Metab. 93, 1046–1053 (2008).
Peschke, E., Bach, A.G. & Muhlbauer, E. Parallel signaling pathways of melatonin in the pancreatic beta-cell. J. Pineal Res. 40, 184–191 (2006).
Peschke, E. et al. Melatonin and type 2 diabetes - a possible link? J. Pineal Res. 42, 350–358 (2007).
Berglund, G. et al. Long-term outcome of the Malmo preventive project: mortality and cardiovascular morbidity. J. Intern. Med. 247, 19–29 (2000).
Lyssenko, V. et al. Clinical risk factors, DNA variants, and the development of type 2 diabetes. N. Engl. J. Med. 359, 2220–2232 (2008).
Lyssenko, V. et al. Genetic prediction of future type 2 diabetes. PLoS Med. 2, e345 (2005).
Lyssenko, V. et al. Predictors of and longitudinal changes in insulin sensitivity & secretion preceding onset of type 2 diabetes. Diabetes 54, 166–174 (2005).
Steil, G.M., Volund, A., Kahn, S.E. & Bergman, R.N. Reduced sample number for calculation of insulin sensitivity and glucose effectiveness from the minimal model. Suitability for use in population studies. Diabetes 42, 250–256 (1993).
Yang, Y.J., Youn, J.H. & Bergman, R.N. Modified protocols improve insulin sensitivity estimation using the minimal model. Am. J. Physiol. 253, E595–E602 (1987).
Bergman, R.N., Ider, Y.Z., Bowden, C.R. & Cobelli, C. Quantitative estimation of insulin sensitivity. Am. J. Physiol. 236, E667–E677 (1979).
Ward, W.K., Bolgiano, D.C., McKnight, B., Halter, J.B. & Porte, D. Jr. Diminished B cell secretory capacity in patients with noninsulin-dependent diabetes mellitus. J. Clin. Invest. 74, 1318–1328 (1984).
Matsuda, M. & DeFronzo, R.A. Insulin sensitivity indices obtained from oral glucose tolerance testing: comparison with the euglycemic insulin clamp. Diabetes Care 22, 1462–1470 (1999).
Hanson, R.L. et al. Evaluation of simple indices of insulin sensitivity and insulin secretion for use in epidemiologic studies. Am. J. Epidemiol. 151, 190–198 (2000).
Wierup, N., Björkqvist, M., Kuhar, M.J., Mulder, H. & Sundler, F. CART regulates islet hormone secretion and is expressed in the beta-cells of type 2 diabetic rats. Diabetes 55, 305–311 (2006).
Del Guerra, S. et al. Functional and molecular defects of pancreatic islets in human type 2 diabetes. Diabetes 54, 727–735 (2005).
Chen, W.M. & Abecasis, G.R. Family-based association tests for genomewide association scans. Am. J. Hum. Genet. 81, 913–926 (2007).
Acknowledgements
The DGI study was supported by a grant from Novartis.
Studies in Malmoe were supported by grants from the Swedish Research Council, including a Linné grant (No. 31475113580), the Diabetes Programme at Lund University, the Påhlsson Foundation, the Heart and Lung Foundation, the Wallenberg Foundation, the Swedish Diabetes Research Society, the Crafoord Foundation, Swedish Medical Society, Swedish Royal Physiographic Society, a Nordic Centre of Excellence Grant in Disease Genetics, the Finnish Diabetes Research Society, the Sigrid Juselius Foundation, Folkhälsan Research Foundation, Novo Nordisk Foundation, the European Network of Genomic and Genetic Epidemiology (ENGAGE), the Wallenberg Foundation, the European Foundation for the Study of Diabetes (EFSD) and the Human Tissue facility at the Lund University Diabetes Center. Studies in human islets were supported in part by the Italian Ministry of University and Research (PRIN 2007-2008) and the European Community (LSHM-CT-2006-518153).
Pancreatic islets at US National Institutes of Health were obtained through the ICR Basic Science Islet Distribution Program (City of Hope Hospital, Joslin Diabetes Center, Northwestern University, Southern California Islet Consortium, University of Alabama Birmingham, University of Illinois, University of Miami, University of Minnesota, University of Pennsylvania, University of Wisconsin and Washington University), the Juvenile Diabetes Research Foundation Islet Resources (Washington University) and the National Disease Resource Interchange (NDRI).
The FUSION study would like to thank the many research volunteers who generously participated in the various studies represented in FUSION. We also thank A.J. Swift, M. Morken, P.S. Chines and N. Narisu for genotying and informatics support. Support for FUSION was provided by the following: NIH grant DK062370 (M. Boehnke), American Diabetes Association research grant 1-05-RA-140 (R.M.W.), DK072193 (K.L. Mohlke) and National Human Genome Research Institute intramural project number 1 Z01 HG000024 (F.S. Collins). The METSIM study was supported by Academy of Finland grant 124243 (M.L.).
Author information
Authors and Affiliations
Contributions
V.L.: DGI GWAS, data analysis and draft of the report. C.L.F.N., M.R.E.: in vitro expression experiments and analysis, and draft of the report. N.W.: immunocytochemistry. A.J.: genotyping and data analysis. P.S.: in vitro expression experiments. M. Bugliani: microarray and human islets experiments. R.S.: DGI GWAS analysis. M.F.: in vitro physiology. N.P.: genotyping. B.I., T.T.: phenotyping in the Botnia study. P.N.: phenotyping in the Malmoe study. J.K.: data analysis in METSIM study. J.T.: phenotyping in the FUSION study. M. Boehnke: PI of the FUSION study. D.A.: PI of the DGI study. F.S.: immunocytochemistry. J.G.E.: phenotyping in the Helsinki Birth Cohort Study. A.U.J.: FUSION GWAS and data analysis. M.L.: PI of the METSIM study. P.M.: microarray and human islets experiments. R.M.W.: FUSION GWAS analysis. H.M.: design and supervision of in vitro study experiments and draft of the report. L.G. designed and supervised all parts of the study and drafted the report. All researchers took part in the revision of the report and approved the final version.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figure 1 (PDF 46 kb)
Rights and permissions
About this article
Cite this article
Lyssenko, V., Nagorny, C., Erdos, M. et al. Common variant in MTNR1B associated with increased risk of type 2 diabetes and impaired early insulin secretion. Nat Genet 41, 82–88 (2009). https://doi.org/10.1038/ng.288
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.288
This article is cited by
-
MTNR1B genotype and effects of carbohydrate quantity and dietary glycaemic index on glycaemic response to an oral glucose load: the OmniCarb trial
Diabetologia (2024)
-
Metabolomic and genetic architecture of gestational diabetes subtypes
Diabetologia (2024)
-
Association of rotating night shift work, CLOCK, MTNR1A, MTNR1B genes polymorphisms and their interactions with type 2 diabetes among steelworkers: a case–control study
BMC Genomics (2023)
-
Association between the MTNR1B, HHEX, SLC30A8, and TCF7L2 single nucleotide polymorphisms and cardiometabolic risk profile in a mixed ancestry South African population
Scientific Reports (2023)
-
Genetics of circadian rhythms and sleep in human health and disease
Nature Reviews Genetics (2023)