Abstract
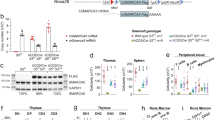
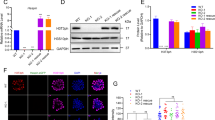
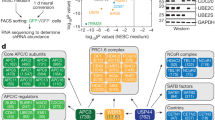
Cell fate can be controlled through asymmetric division and segregation of protein determinants, but the regulation of this process in the hematopoietic system is poorly understood. Here we show that the dynein-binding protein Lis1 is critically required for hematopoietic stem cell function and leukemogenesis. Conditional deletion of Lis1 (also known as Pafah1b1) in the hematopoietic system led to a severe bloodless phenotype, depletion of the stem cell pool and embryonic lethality. Further, real-time imaging revealed that loss of Lis1 caused defects in spindle positioning and inheritance of cell fate determinants, triggering accelerated differentiation. Finally, deletion of Lis1 blocked the propagation of myeloid leukemia and led to a marked improvement in survival, suggesting that Lis1 is also required for oncogenic growth. These data identify a key role for Lis1 in hematopoietic stem cells and mark its directed control of asymmetric division as a critical regulator of normal and malignant hematopoietic development.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
Primary accessions
ArrayExpress
Referenced accessions
NCBI Reference Sequence
References
Siller, K.H. & Doe, C.Q. Lis1/dynactin regulates metaphase spindle orientation in Drosophila neuroblasts. Dev. Biol. 319, 1–9 (2008).
Yingling, J. et al. Neuroepithelial stem cell proliferation requires LIS1 for precise spindle orientation and symmetric division. Cell 132, 474–486 (2008).
Suda, T., Suda, J. & Ogawa, M. Disparate differentiation in mouse hemopoietic colonies derived from paired progenitors. Proc. Natl. Acad. Sci. USA 81, 2520–2524 (1984).
Ting, S.B. et al. Asymmetric segregation and self-renewal of hematopoietic stem and progenitor cells with endocytic Ap2a2. Blood 119, 2510–2522 (2012).
Wu, M. et al. Imaging hematopoietic precursor division in real time. Cell Stem Cell 1, 541–554 (2007).
Hope, K.J. et al. An RNAi screen identifies Msi2 and Prox1 as having opposite roles in the regulation of hematopoietic stem cell activity. Cell Stem Cell 7, 101–113 (2010).
Ito, K. et al. Regulation of reactive oxygen species by Atm is essential for proper response to DNA double-strand breaks in lymphocytes. J. Immunol. 178, 103–110 (2007).
Kharas, M.G. et al. Musashi-2 regulates normal hematopoiesis and promotes aggressive myeloid leukemia. Nat. Med. 16, 903–908 (2010).
de Andrés-Aguayo, L. et al. Musashi 2 is a regulator of the HSC compartment identified by a retroviral insertion screen and knockout mice. Blood 118, 554–564 (2011).
Ito, T. et al. Regulation of myeloid leukaemia by the cell-fate determinant Musashi. Nature 466, 765–768 (2010).
Hirotsune, S. et al. Graded reduction of Pafah1b1 (Lis1) activity results in neuronal migration defects and early embryonic lethality. Nat. Genet. 19, 333–339 (1998).
de Boer, J. et al. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur. J. Immunol. 33, 314–325 (2003).
Almarza, E. et al. Regulatory elements of the vav gene drive transgene expression in hematopoietic stem cells from adult mice. Exp. Hematol. 32, 360–364 (2004).
Ogilvy, S. et al. Promoter elements of vav drive transgene expression in vivo throughout the hematopoietic compartment. Blood 94, 1855–1863 (1999).
Ventura, A. et al. Restoration of p53 function leads to tumour regression in vivo. Nature 445, 661–665 (2007).
Wong, D.J. et al. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell 2, 333–344 (2008).
Venezia, T.A. et al. Molecular signatures of proliferation and quiescence in hematopoietic stem cells. PLoS Biol. 2, e301 (2004).
Eppert, K. et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat. Med. 17, 1086–1093 (2011).
Toyoshima, F. & Nishida, E. Integrin-mediated adhesion orients the spindle parallel to the substratum in an EB1- and myosin X–dependent manner. EMBO J. 26, 1487–1498 (2007).
Kanda, T., Sullivan, K.F. & Wahl, G.M. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr. Biol. 8, 377–385 (1998).
Day, D. et al. A method for prolonged imaging of motile lymphocytes. Immunol. Cell Biol. 87, 154–158 (2009).
Feng, Y. & Walsh, C.A. Mitotic spindle regulation by Nde1 controls cerebral cortical size. Neuron 44, 279–293 (2004).
Reya, T., Morrison, S.J., Clarke, M.F. & Weissman, I.L. Stem cells, cancer, and cancer stem cells. Nature 414, 105–111 (2001).
Dash, A.B. et al. A murine model of CML blast crisis induced by cooperation between BCR/ABL and NUP98/HOXA9. Proc. Natl. Acad. Sci. USA 99, 7622–7627 (2002).
Mayotte, N., Roy, D.C., Yao, J., Kroon, E. & Sauvageau, G. Oncogenic interaction between BCR-ABL and NUP98-HOXA9 demonstrated by the use of an in vitro purging culture system. Blood 100, 4177–4184 (2002).
Neering, S.J. et al. Leukemia stem cells in a genetically defined murine model of blast-crisis CML. Blood 110, 2578–2585 (2007).
Zuber, J. et al. Mouse models of human AML accurately predict chemotherapy response. Genes Dev. 23, 877–889 (2009).
Tsai, F.Y. et al. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature 371, 221–226 (1994).
Wang, Q. et al. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc. Natl. Acad. Sci. USA 93, 3444–3449 (1996).
Okuda, T., van Deursen, J., Hiebert, S.W., Grosveld, G. & Downing, J.R. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell 84, 321–330 (1996).
Porcher, C. et al. The T cell leukemia oncoprotein SCL/tal-1 is essential for development of all hematopoietic lineages. Cell 86, 47–57 (1996).
Gan, B. et al. Lkb1 regulates quiescence and metabolic homeostasis of haematopoietic stem cells. Nature 468, 701–704 (2010).
Nakada, D., Saunders, T.L. & Morrison, S.J. Lkb1 regulates cell cycle and energy metabolism in haematopoietic stem cells. Nature 468, 653–658 (2010).
Bonaccorsi, S. et al. The Drosophila Lkb1 kinase is required for spindle formation and asymmetric neuroblast division. Development 134, 2183–2193 (2007).
Calabretta, B. & Perrotti, D. The biology of CML blast crisis. Blood 103, 4010–4022 (2004).
Stingl, J. & Caldas, C. Molecular heterogeneity of breast carcinomas and the cancer stem cell hypothesis. Nat. Rev. Cancer 7, 791–799 (2007).
Maher, E.A. et al. Malignant glioma: genetics and biology of a grave matter. Genes Dev. 15, 1311–1333 (2001).
Pece, S. et al. Loss of negative regulation by Numb over Notch is relevant to human breast carcinogenesis. J. Cell Biol. 167, 215–221 (2004).
Bello, B., Reichert, H. & Hirth, F. The brain tumor gene negatively regulates neural progenitor cell proliferation in the larval central brain of Drosophila. Development 133, 2639–2648 (2006).
Betschinger, J., Mechtler, K. & Knoblich, J.A. Asymmetric segregation of the tumor suppressor Brat regulates self-renewal in Drosophila neural stem cells. Cell 124, 1241–1253 (2006).
Caussinus, E. & Gonzalez, C. Induction of tumor growth by altered stem-cell asymmetric division in Drosophila melanogaster. Nat. Genet. 37, 1125–1129 (2005).
Lee, C.Y. et al. Drosophila Aurora-A kinase inhibits neuroblast self-renewal by regulating aPKC/Numb cortical polarity and spindle orientation. Genes Dev. 20, 3464–3474 (2006).
Lee, C.Y., Wilkinson, B.D., Siegrist, S.E., Wharton, R.P. & Doe, C.Q. Brat is a Miranda cargo protein that promotes neuronal differentiation and inhibits neuroblast self-renewal. Dev. Cell 10, 441–449 (2006).
Wang, H. et al. Aurora-A acts as a tumor suppressor and regulates self-renewal of Drosophila neuroblasts. Genes Dev. 20, 3453–3463 (2006).
Yang, Z.J. et al. Medulloblastoma can be initiated by deletion of Patched in lineage-restricted progenitors or stem cells. Cancer Cell 14, 135–145 (2008).
Domen, J., Cheshier, S.H. & Weissman, I.L. The role of apoptosis in the regulation of hematopoietic stem cells: overexpression of Bcl-2 increases both their number and repopulation potential. J. Exp. Med. 191, 253–264 (2000).
Qin, X.F., An, D.S., Chen, I.S. & Baltimore, D. Inhibiting HIV-1 infection in human T cells by lentiviral-mediated delivery of small interfering RNA against CCR5. Proc. Natl. Acad. Sci. USA 100, 183–188 (2003).
Sásik, R., Woelk, C.H. & Corbeil, J. Microarray truths and consequences. J. Mol. Endocrinol. 33, 1–9 (2004).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 (1995).
Tusher, V.G., Tibshirani, R. & Chu, G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98, 5116–5121 (2001).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550 (2005).
Arnold, B.C., Balakrishnan, N. & Nagaraja, H.N. A First Course in Order Statistics (Wiley Series in Probability and Statistics) (John Wiley & Sons, New York, 1992).
Metzeler, K.H. et al. An 86-probe-set gene-expression signature predicts survival in cytogenetically normal acute myeloid leukemia. Blood 112, 4193–4201 (2008).
Somervaille, T.C. et al. Hierarchical maintenance of MLL myeloid leukemia stem cells employs a transcriptional program shared with embryonic rather than adult stem cells. Cell Stem Cell 4, 129–140 (2009).
Yagi, T. et al. Identification of a gene expression signature associated with pediatric AML prognosis. Blood 102, 1849–1856 (2003).
Acknowledgements
We are grateful to B. Hogan, J. Chang, A. Desai, J. Gleeson, J.E. Lee, M.F. Wu, M. Sander and J. Koop for experimental advice and reagents. We would also like to thank M. Kritzik for advice and comments on the manuscript; M. Nakamura for experimental help; M. Cook, L. Matinek, B. Harvat, E. O'Conner and K. Marquez for cell sorting; and W. Pear (University of Pennsylvania) and A.M. Pendergast (Duke University) for the BCR-ABL construct, D.G. Gilliland (University of Pennsylvania) for the NUP98-HOXA9 construct, S. Armstrong (Memorial Sloan-Kettering Cancer Center) for the MLL-AF9 construct, C. Counter (Duke University) for NRASG12V, S. Russell (Peter MacCallum Cancer Centre) for the mCherry–α-tubulin construct, G. Wahl (Salk Institute) for the H2B-GFP (pEGFPN1) vector and D. Kioussis (Medical Research Council National Institute for Medical Research) for the Vav1-cre transgenic line. B.Z. and C.S.K. received support from US National Institutes of Health (NIH) Cancer Biology Training Grant (T32 CA 59365-18) and NIH Pharmacological Sciences Training Program (T32 GM007752), respectively. T.I. is a recipient of a California Institute for Regenerative Medicine interdisciplinary stem cell training program fellowship, and T.K. is supported by a postdoctoral fellowship from the Japanese Society for the Promotion of Science. This work was also supported by a Leukemia and Lymphoma Society Scholar Award, the University of California San Diego Moores Cancer Center National Cancer Institute Core Grant, P30CA23100, as well as by NIH grants DK63031, HL097767 and DP1 CA174422 awarded to T.R.
Author information
Authors and Affiliations
Contributions
B.Z. and T.I. planned and designed the research, performed the majority of experiments and helped write the manuscript. B.Z. and J.B. developed all real-time imaging methods for visualizing and tracking spindle orientation in primary hematopoietic cells. A.B., J.B., T.K., J.W., C.S.K., H.Y.K. and O.A. provided experimental data and help. D.R., H.E.B., C.C. and V.G.O. provided primary patient samples and experimental advice. R.S. and G.H. carried out all bioinformatics analysis on microarray data. T.R. planned and guided the project, provided experimental advice and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–20, Supplementary Tables 1 and 2. (PDF 32604 kb)
Imaging Cell Division in Real Time
Cells were co-infected with H2B-GFP and mCherry-α-tubulin fusion constructs and imaged over time. Representative movies show A. HeLa cell, B. M1 Cell, and C. Primary Hematopoietic Stem & Progenitor Cells undergoing cell division (H2B-GFP is shown in green and α-tubulin is shown in magenta). (MOV 4040 kb)
Symmetric inheritance of Numb
HSC-enriched cells (KLS) were co-infected with Numb-CFP and mCherry-α-tubulin fusion constructs and imaged over time. Representative movie shows a KLS cell undergoing symmetric division (Numb is shown in green and α-tubulin is shown in red). (MOV 15036 kb)
Asymmetric inheritance of Numb
HSC-enriched cells (KLS) were co-infected with Numb-CFP and mCherry-α-tubulin fusion constructs and imaged over time. Representative movie shows a KLS cell undergoing asymmetric division (Numb is shown in green and α-tubulin is shown in red). (MOV 16403 kb)
Asymmetric inheritance of Numb
HSC-enriched cells (KLS) were co-infected with Numb-YFP and mCherry-α-tubulin fusion constructs and imaged over time. Representative movie shows a KLS cell undergoing asymmetric division (Numb is shown in green and α-tubulin is shown in red). (MOV 12302 kb)
Rights and permissions
About this article
Cite this article
Zimdahl, B., Ito, T., Blevins, A. et al. Lis1 regulates asymmetric division in hematopoietic stem cells and in leukemia. Nat Genet 46, 245–252 (2014). https://doi.org/10.1038/ng.2889
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2889
This article is cited by
-
Spindle positioning and its impact on vertebrate tissue architecture and cell fate
Nature Reviews Molecular Cell Biology (2021)
-
LIGHT/LTβR signaling regulates self-renewal and differentiation of hematopoietic and leukemia stem cells
Nature Communications (2021)
-
AMD1 is required for the maintenance of leukemic stem cells and promotes chronic myeloid leukemic growth
Oncogene (2021)
-
Hematopoiesis under telomere attrition at the single-cell resolution
Nature Communications (2021)
-
An in vivo genome-wide CRISPR screen identifies the RNA-binding protein Staufen2 as a key regulator of myeloid leukemia
Nature Cancer (2020)