Abstract
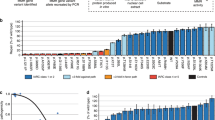
The clinical classification of hereditary sequence variants identified in disease-related genes directly affects clinical management of patients and their relatives. The International Society for Gastrointestinal Hereditary Tumours (InSiGHT) undertook a collaborative effort to develop, test and apply a standardized classification scheme to constitutional variants in the Lynch syndrome–associated genes MLH1, MSH2, MSH6 and PMS2. Unpublished data submission was encouraged to assist in variant classification and was recognized through microattribution. The scheme was refined by multidisciplinary expert committee review of the clinical and functional data available for variants, applied to 2,360 sequence alterations, and disseminated online. Assessment using validated criteria altered classifications for 66% of 12,006 database entries. Clinical recommendations based on transparent evaluation are now possible for 1,370 variants that were not obviously protein truncating from nomenclature. This large-scale endeavor will facilitate the consistent management of families suspected to have Lynch syndrome and demonstrates the value of multidisciplinary collaboration in the curation and classification of variants in public locus-specific databases.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Vasen, H.F. et al. Revised guidelines for the clinical management of Lynch syndrome (HNPCC): recommendations by a group of European experts. Gut 62, 812–823 (2013).
Umar, A. et al. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J. Natl. Cancer Inst. 96, 261–268 (2004).
van Oers, J.M. et al. PMS2 endonuclease activity has distinct biological functions and is essential for genome maintenance. Proc. Natl. Acad. Sci. USA 107, 13384–13389 (2010).
Win, A.K. et al. Risks of primary extracolonic cancers following colorectal cancer in lynch syndrome. J. Natl. Cancer Inst. 104, 1363–1372 (2012).
Buerki, N. et al. Evidence for breast cancer as an integral part of Lynch syndrome. Genes Chromosom. Cancer 51, 83–91 (2012).
Scott, R.J. et al. Hereditary nonpolyposis colorectal cancer in 95 families: differences and similarities between mutation-positive and mutation-negative kindreds. Am. J. Hum. Genet. 68, 118–127 (2001).
Grindedal, E.M. et al. Germ-line mutations in mismatch repair genes associated with prostate cancer. Cancer Epidemiol. Biomarkers Prev. 18, 2460–2467 (2009).
Win, A.K. et al. Colorectal and other cancer risks for carriers and noncarriers from families with a DNA mismatch repair gene mutation: a prospective cohort study. J. Clin. Oncol. 30, 958–964 (2012).
Järvinen, H.J. et al. Ten years after mutation testing for Lynch syndrome: cancer incidence and outcome in mutation-positive and mutation-negative family members. J. Clin. Oncol. 27, 4793–4797 (2009).
Plon, S.E. et al. Sequence variant classification and reporting: recommendations for improving the interpretation of cancer susceptibility genetic test results. Hum. Mutat. 29, 1282–1291 (2008).
Tavtigian, S.V., Greenblatt, M.S., Goldgar, D.E. & Boffetta, P. Assessing pathogenicity: overview of results from the IARC Unclassified Genetic Variants Working Group. Hum. Mutat. 29, 1261–1264 (2008).
Richards, C.S. et al. ACMG recommendations for standards for interpretation and reporting of sequence variations: Revisions 2007. Genet. Med. 10, 294–300 (2008).
Easton, D.F. et al. A systematic genetic assessment of 1,433 sequence variants of unknown clinical significance in the BRCA1 and BRCA2 breast cancer–predisposition genes. Am. J. Hum. Genet. 81, 873–883 (2007).
Goldgar, D.E. et al. Genetic evidence and integration of various data sources for classifying uncertain variants into a single model. Hum. Mutat. 29, 1265–1272 (2008).
Goldgar, D.E. et al. Integrated evaluation of DNA sequence variants of unknown clinical significance: application to BRCA1 and BRCA2. Am. J. Hum. Genet. 75, 535–544 (2004).
Thompson, B.A. et al. A multifactorial likelihood model for MMR gene variant classification incorporating probabilities based on sequence bioinformatics and tumor characteristics: a report from the Colon Cancer Family Registry. Hum. Mutat. 34, 200–209 (2013).
Spurdle, A.B., Couch, F.J., Hogervorst, F.B., Radice, P. & Sinilnikova, O.M. Prediction and assessment of splicing alterations: implications for clinical testing. Hum. Mutat. 29, 1304–1313 (2008).
Greenblatt, M.S. et al. Locus-specific databases and recommendations to strengthen their contribution to the classification of variants in cancer susceptibility genes. Hum. Mutat. 29, 1273–1281 (2008).
Plazzer, J.P. et al. The InSiGHT database: utilizing 100 years of insights into Lynch syndrome. Fam. Cancer 12, 175–180 (2013).
Peltomäki, P. & Vasen, H. Mutations associated with HNPCC predisposition—update of ICG-HNPCC/INSiGHT mutation database. Dis. Markers 20, 269–276 (2004).
Peltomäki, P. & Vasen, H.F. Mutations predisposing to hereditary nonpolyposis colorectal cancer: database and results of a collaborative study. The International Collaborative Group on Hereditary Nonpolyposis Colorectal Cancer. Gastroenterology 113, 1146–1158 (1997).
Ou, J. et al. Functional analysis helps to clarify the clinical importance of unclassified variants in DNA mismatch repair genes. Hum. Mutat. 28, 1047–1054 (2007).
Woods, M.O. et al. A new variant database for mismatch repair genes associated with Lynch syndrome. Hum. Mutat. 28, 669–673 (2007).
Giardine, B. et al. Systematic documentation and analysis of human genetic variation in hemoglobinopathies using the microattribution approach. Nat. Genet. 43, 295–301 (2011).
Fox, B.I. et al. Developing an expert panel process to refine health outcome definitions in observational data. J. Biomed. Inform. 46, 795–804 (2013).
Kohonen-Corish, M.R. et al. Deciphering the colon cancer genes—report of the InSiGHT–Human Variome Project Workshop, UNESCO, Paris 2010. Hum. Mutat. 32, 491–494 (2011).
Thompson, D., Easton, D.F. & Goldgar, D.E. A full-likelihood method for the evaluation of causality of sequence variants from family data. Am. J. Hum. Genet. 73, 652–655 (2003).
Senter, L. et al. The clinical phenotype of Lynch syndrome due to germ-line PMS2 mutations. Gastroenterology 135, 419–428 (2008).
Baglietto, L. et al. Risks of Lynch syndrome cancers for MSH6 mutation carriers. J. Natl. Cancer Inst. 102, 193–201 (2010).
Bonadona, V. et al. Cancer risks associated with germline mutations in MLH1, MSH2, and MSH6 genes in Lynch syndrome. J. Am. Med. Assoc. 305, 2304–2310 (2011).
Mangold, E. et al. Spectrum and frequencies of mutations in MSH2 and MLH1 identified in 1,721 German families suspected of hereditary nonpolyposis colorectal cancer. Int. J. Cancer 116, 692–702 (2005).
Barnetson, R.A. et al. Identification and survival of carriers of mutations in DNA mismatch-repair genes in colon cancer. N. Engl. J. Med. 354, 2751–2763 (2006).
Patrinos, G.P. et al. Microattribution and nanopublication as means to incentivize the placement of human genome variation data into the public domain. Hum. Mutat. 33, 1503–1512 (2012).
Thompson, B.A. et al. Calibration of multiple in silico tools for predicting pathogenicity of mismatch repair gene missense substitutions. Hum. Mutat. 34, 255–265 (2013).
Vallée, M.P. et al. Classification of missense substitutions in the BRCA genes: a database dedicated to Ex-UVs. Hum. Mutat. 33, 22–28 (2012).
Drost, M. et al. A rapid and cell-free assay to test the activity of Lynch syndrome–associated MSH2 and MSH6 missense variants. Hum. Mutat. 33, 488–494 (2012).
Heinen, C.D. & Juel Rasmussen, L. Determining the functional significance of mismatch repair gene missense variants using biochemical and cellular assays. Hered. Cancer Clin. Pract. 10, 9 (2012).
Couch, F.J. et al. Assessment of functional effects of unclassified genetic variants. Hum. Mutat. 29, 1314–1326 (2008).
Rasmussen, L.J. et al. Pathological assessment of mismatch repair gene variants in Lynch syndrome: past, present and future. Hum. Mutat. 33, 1617–1625 (2012).
Leenen, C.H. et al. Pitfalls in molecular analysis for mismatch repair deficiency in a family with biallelic PMS2 germline mutations. Clin. Genet. 80, 558–565 (2011).
Mead, L.J. et al. Microsatellite instability markers for identifying early-onset colorectal cancers caused by germ-line mutations in DNA mismatch repair genes. Clin. Cancer Res. 13, 2865–2869 (2007).
Plaschke, J. et al. Lower incidence of colorectal cancer and later age of disease onset in 27 families with pathogenic MSH6 germline mutations compared with families with MLH1 or MSH2 mutations: the German Hereditary Nonpolyposis Colorectal Cancer Consortium. J. Clin. Oncol. 22, 4486–4494 (2004).
Wu, Y. et al. Association of hereditary nonpolyposis colorectal cancer–related tumors displaying low microsatellite instability with MSH6 germline mutations. Am. J. Hum. Genet. 65, 1291–1298 (1999).
You, J.F. et al. Tumours with loss of MSH6 expression are MSI-H when screened with a pentaplex of five mononucleotide repeats. Br. J. Cancer 103, 1840–1845 (2010).
Spurdle, A.B. et al. BRCA1 R1699Q variant displaying ambiguous functional abrogation confers intermediate breast and ovarian cancer risk. J. Med. Genet. 49, 525–532 (2012).
Xie, J. et al. An MLH1 mutation links BACH1/FANCJ to colon cancer, signaling, and insight toward directed therapy. Cancer Prev. Res. (Phila.) 3, 1409–1416 (2010).
Kosinski, J., Hinrichsen, I., Bujnicki, J.M., Friedhoff, P. & Plotz, G. Identification of Lynch syndrome mutations in the MLH1-PMS2 interface that disturb dimerization and mismatch repair. Hum. Mutat. 31, 975–982 (2010).
Takahashi, M. et al. Functional analysis of human MLH1 variants using yeast and in vitro mismatch repair assays. Cancer Res. 67, 4595–4604 (2007).
Hinrichsen, I. et al. Expression defect size among unclassified MLH1 variants determines pathogenicity in Lynch syndrome diagnosis. Clin. Cancer Res. 19, 2432–2441 (2013).
Wildeman, M., van Ophuizen, E., den Dunnen, J.T. & Taschner, P.E. Improving sequence variant descriptions in mutation databases and literature using the Mutalyzer sequence variation nomenclature checker. Hum. Mutat. 29, 6–13 (2008).
Arnold, S. et al. Classifying MLH1 and MSH2 variants using bioinformatic prediction, splicing assays, segregation, and tumor characteristics. Hum. Mutat. 30, 757–770 (2009).
Spurdle, A.B. Clinical relevance of rare germline sequence variants in cancer genes: evolution and application of classification models. Curr. Opin. Genet. Dev. 20, 315–323 (2010).
Wimmer, K. & Etzler, J. Constitutional mismatch repair–deficiency syndrome: have we so far seen only the tip of an iceberg? Hum. Genet. 124, 105–122 (2008).
1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073 (2010).
ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Acknowledgements
We thank all submitters of data to the InSiGHT database (retrospective and prospective), the Colon Cancer Family Registry and the German Hereditary Non-polyposis Colorectal Cancer Consortium for their contributions of unpublished data, acknowledged formally through microattribution. We would also like to acknowledge L. Marquart for providing statistical advice and T. O'Mara for providing advice and assistance with the statistical package R. We are extremely grateful to the Hicks Foundation (Australia) for inaugural support of InSiGHT database curator J.-P.P. Funding for VIC teleconferences was provided by the Cancer Council of Victoria. B.A.T. is supported by a Cancer Council of Queensland PhD scholarship and by a Queensland Institute of Medical Research PhD Top-Up award. A.B.S. is a National Health and Medical Research Council Senior Research Fellow. The work performed by A.B.S. and B.A.T. was additionally supported by Cancer Australia (1010859). M.G. is supported by a grant from the Tuscan Tumor Institute (ITT). J.-P.P. is currently supported by the Royal Melbourne Hospital Foundation. S.V.T., M.S.G., A.B.S., L.J.R. and R.S. are supported by grant 1R01CA164944 from the National Cancer Institute/US National Institutes of Health (NCI/US NIH). G.C. and M.P. were supported by the Ministerio de Ciencia e Innovación (SAF 12-33636) and by the Fundación Científica de la Asociación Española Contra el Cáncer. A.F. is supported by the French National Cancer Institute and by the Institut National du Cancer (INCa) French MMR Committee. S.M.F. is supported by grants from the Association of International Cancer Research (12-1087) and by the Medical Research Council UK (MR/K018647/1). NHS Wales National Institute for Health and Social Care (NIHSCR) funding was provided to I.M.F. via the Cardiff & Vale University Health Board. D.E.G. is supported by funding from Mayo Specialized Programs of Research Excellence (SPORE) grant P50CA11620106 (principal investigator J. Ingle). C.D.H. is funded by US NIH grant R01 CA115783-02/CA/NCI. E.H.-F. and M.M. are supported by German Cancer Aid (Deutsche Krebshilfe) and by the Wilhelm Sander Foundation. M.K.-C. is funded by Cancer Institute NSW. S.Y.L. is supported by the Hong Kong Cancer Fund. A.M. is supported by the French National Cancer Institute and by the Direction Générale de l'Offre des Soins (INCa/DGOS). The Sigrid Juselius Foundation funds M.N. Funding for P.P. is provided by the European Research Council (FP7-ERC-232635). L.J.R. is funded by Nordea-Fonden. B.R.-P. is supported by German Cancer Aid. M.O.W. was supported by the Canadian Cancer Society Research Institute (grant 18223).
Author information
Authors and Affiliations
Consortia
Contributions
A.B.S. and B.A.T. drafted the manuscript. B.A.T. conducted InSiGHT database nomenclature standardization and data cleaning, systematic literature and data review, statistical analyses and final data analyses and assisted in the presentation of data in web-based format. B.A.T., A.B.S., S.V.T., M.S.G., D.E.G. and M.G. formulated the baseline guidelines for consideration by VIC members. B.A.T. and A.B.S. developed the functional flowchart and, with L.J.R., C.D.H., G.C., M.P., A.M., B.R.-P., E.H.-F., M.S.G., M.M., T.F. and M.N. formed the functional subcommittee contributing to the supporting documents for functional assay interpretation. D.E.G. provided statistical input. J.-P.P. provided data management, organized teleconferences, collated information after teleconferences, coordinated microattribution and was responsible for the presentation of data in web-based format. J.T.d.D. provided support for the LOVD database and created the LOVD nanopublications. F.M. is the responsible InSiGHT Councilor who initiated the concept of VIC in 2007 and has been responsible for advocating for funding and organizing the face-to-face meeting in Paris. M.G. coordinated VIC and chaired teleconferences and face-to-face meetings. B.A.T., A.B.S., S.V.T., M.S.G., D.E.G., M.G., F.M., L.J.R., C.D.H., G.C., M.P., A.M., B.R.-P., E.H.-F., M.S.G., M.M., T.F., M.N., K.A., F.A.-M., B.B., I.B., D.d.S., A.F., M.P.F., S.M.F., I.M.F., M.K.-C., K.L.R., S.Y.L., P.M., P.P., M.Q., R.R., R.J.S., R.S., C.M.T., T.W., J.W. and M.O.W. provided critique of the classification criteria and/or participated in review of variants at teleconferences or face-to-face meetings or by e-mail and provided critical review of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
A full list of collaborators assigned microattributions for this study appears at the end of the paper with affiliations.
Supplementary information
Supplementary Text and Figures
Supplementary Note, Supplementary Figures 1 and 2, and Supplementary Tables 2–7 and 9 (PDF 4174 kb)
Supplementary Table 1
Details of non-constitutional variants on the InSiGHT database (XLSX 31 kb)
Supplementary Table 8
Summary justifications for class 4: likely pathogenic and class 5: pathogenic “not obviously truncating” nonsynonymous variants classified by the InSiGHT Variant Interpretation Committee (XLSX 67 kb)
Rights and permissions
About this article
Cite this article
Thompson, B., Spurdle, A., Plazzer, JP. et al. Application of a 5-tiered scheme for standardized classification of 2,360 unique mismatch repair gene variants in the InSiGHT locus-specific database. Nat Genet 46, 107–115 (2014). https://doi.org/10.1038/ng.2854
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2854
This article is cited by
-
Lynch syndrome, molecular mechanisms and variant classification
British Journal of Cancer (2023)
-
Hereditäre kolorektale Karzinogenese
Die Pathologie (2023)
-
Splicing analyses for variants in MMR genes: best practice recommendations from the European Mismatch Repair Working Group
European Journal of Human Genetics (2022)
-
Clinical challenges in interpreting multiple pathogenic mutations in single patients
Hereditary Cancer in Clinical Practice (2021)
-
Room for improvement: One third of Lynch syndrome patients presenting for genetic testing in a highly specialised centre in Stockholm already have cancer
Hereditary Cancer in Clinical Practice (2021)