Abstract
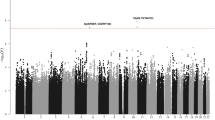
Genome-wide association studies (GWAS) of chronic lymphocytic leukemia (CLL) have shown that common genetic variation contributes to the heritable risk of CLL. To identify additional CLL susceptibility loci, we conducted a GWAS and performed a meta-analysis with a published GWAS totaling 1,739 individuals with CLL (cases) and 5,199 controls with validation in an additional 1,144 cases and 3,151 controls. A combined analysis identified new susceptibility loci mapping to 3q26.2 (rs10936599, P = 1.74 × 10−9), 4q26 (rs6858698, P = 3.07 × 10−9), 6q25.2 (IPCEF1, rs2236256, P = 1.50 × 10−10) and 7q31.33 (POT1, rs17246404, P = 3.40 × 10−8). Additionally, we identified a promising association at 5p15.33 (CLPTM1L, rs31490, P = 1.72 × 10−7) and validated recently reported putative associations at 5p15.33 (TERT, rs10069690, P = 1.12 × 10−10) and 8q22.3 (rs2511714, P = 2.90 × 10−9). These findings provide further insights into the genetic and biological basis of inherited genetic susceptibility to CLL.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Stevenson, F.K. & Caligaris-Cappio, F. Chronic lymphocytic leukemia: revelations from the B-cell receptor. Blood 103, 4389–4395 (2004).
Goldin, L.R., Bjorkholm, M., Kristinsson, S.Y., Turesson, I. & Landgren, O. Elevated risk of chronic lymphocytic leukemia and other indolent non-Hodgkin's lymphomas among relatives of patients with chronic lymphocytic leukemia. Haematologica 94, 647–653 (2009).
Di Bernardo, M.C. et al. A genome-wide association study identifies six susceptibility loci for chronic lymphocytic leukemia. Nat. Genet. 40, 1204–1210 (2008).
Crowther-Swanepoel, D. et al. Common variants at 2q37.3, 8q24.21, 15q21.3 and 16q24.1 influence chronic lymphocytic leukemia risk. Nat. Genet. 42, 132–136 (2010).
Slager, S.L. et al. Common variation at 6p21.31 (BAK1) influences the risk of chronic lymphocytic leukemia. Blood 120, 843–846 (2012).
Berndt, S.I. et al. Genome-wide association study identifies multiple risk loci for chronic lymphocytic leukemia. Nat. Genet. 45, 868–876 (2013).
Ernst, J. & Kellis, M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat. Biotechnol. 28, 817–825 (2010).
Houlston, R.S. et al. Meta-analysis of three genome-wide association studies identifies susceptibility loci for colorectal cancer at 1q41, 3q26.2, 12q13.13 and 20q13.33. Nat. Genet. 42, 973–977 (2010).
Chubb, D. et al. Common variation at 3q26.2, 6p21.33, 17p11.2 and 22q13.1 influences multiple myeloma risk. Nat. Genet. 45, 1221–1225 (2013).
Jones, A.M. et al. TERC polymorphisms are associated both with susceptibility to colorectal cancer and with longer telomeres. Gut 61, 248–254 (2012).
Codd, V. et al. Identification of seven loci affecting mean telomere length and their association with disease. Nat. Genet. 45, 422–427 (2013).
Verma-Gaur, J., Hauser, J. & Grundstrom, T. Negative feedback regulation of antigen receptors through calmodulin inhibition of E2A. J. Immunol. 188, 6175–6183 (2012).
Ramsay, A.J. et al. POT1 mutations cause telomere dysfunction in chronic lymphocytic leukemia. Nat. Genet. 45, 526–530 (2013).
Petersen, G.M. et al. A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat. Genet. 42, 224–228 (2010).
Barrett, J.H. et al. Genome-wide association study identifies three new melanoma susceptibility loci. Nat. Genet. 43, 1108–1113 (2011).
Stacey, S.N. et al. New common variants affecting susceptibility to basal cell carcinoma. Nat. Genet. 41, 909–914 (2009).
Wang, Y. et al. Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat. Genet. 40, 1407–1409 (2008).
Rothman, N. et al. A multi-stage genome-wide association study of bladder cancer identifies multiple susceptibility loci. Nat. Genet. 42, 978–984 (2010).
Bojesen, S.E. et al. Multiple independent variants at the TERT locus are associated with telomere length and risks of breast and ovarian cancer. Nat. Genet. 45, 371–384 (2013).
Haiman, C.A. et al. A common variant at the TERT-CLPTM1L locus is associated with estrogen receptor–negative breast cancer. Nat. Genet. 43, 1210–1214 (2011).
Karlsson, R. et al. Investigation of six testicular germ cell tumor susceptibility genes suggests a parent-of-origin effect in SPRY4. Hum. Mol. Genet. 22, 3373–3380 (2013).
Hamblin, T.J., Davis, Z., Gardiner, A., Oscier, D.G. & Stevenson, F.K. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood 94, 1848–1854 (1999).
Grundberg, E. et al. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat. Genet. 44, 1084–1089 (2012).
Di Bernardo, M.C., Broderick, P., Catovsky, D. & Houlston, R.S. Common genetic variation contributes significantly to the risk of developing chronic lymphocytic leukemia. Haematologica 98, e23–e24 (2013).
Swerdlow, S.H. et al. World Health Organization Classification of Tumours of Haematopoietic and Lymphoid Tissues (IARC Press, Lyon, France, 2008).
Catovsky, D. et al. Assessment of fludarabine plus cyclophosphamide for patients with chronic lymphocytic leukaemia (the LRF CLL4 Trial): a randomised controlled trial. Lancet 370, 230–239 (2007).
Power, C. & Elliott, J. Cohort profile: 1958 British birth cohort (National Child Development Study). Int. J. Epidemiol. 35, 34–41 (2006).
Penegar, S. et al. National study of colorectal cancer genetics. Br. J. Cancer 97, 1305–1309 (2007).
Eisen, T., Matakidou, A. & Houlston, R. Identification of low penetrance alleles for lung cancer: the GEnetic Lung CAncer Predisposition Study (GELCAPS). BMC Cancer 8, 244 (2008).
Smedby, K.E. et al. Ultraviolet radiation exposure and risk of malignant lymphomas. J. Natl. Cancer Inst. 97, 199–209 (2005).
van Dongen, J.J. et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia 17, 2257–2317 (2003).
van Krieken, J.H. et al. Improved reliability of lymphoma diagnostics via PCR-based clonality testing: report of the BIOMED-2 Concerted Action BHM4-CT98-3936. Leukemia 21, 201–206 (2007).
Cawthon, R.M. Telomere measurement by quantitative PCR. Nucleic Acids Res. 30, e47 (2002).
Nordfjäll, K., Osterman, P., Melander, O., Nilsson, P. & Roos, G. hTERT (–1327)T/C polymorphism is not associated with age-related telomere attrition in peripheral blood. Biochem. Biophys. Res. Commun. 358, 215–218 (2007).
Mansouri, L. et al. Short telomere length is associated with NOTCH1/SF3B1/TP53 aberrations and poor outcome in newly diagnosed chronic lymphocytic leukemia patients. Am. J. Hematol. 88, 647–651 (2013).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Clayton, D.G. et al. Population structure, differential bias and genomic control in a large-scale, case-control association study. Nat. Genet. 37, 1243–1246 (2005).
Petitti, D. Meta-analysis, Decision Analysis, and Cost-effectiveness Analysis (Oxford University Press, 1994).
Higgins, J.P. & Thompson, S.G. Quantifying heterogeneity in a meta-analysis. Stat. Med. 21, 1539–1558 (2002).
Myers, S., Bottolo, L., Freeman, C., McVean, G. & Donnelly, P. A fine-scale map of recombination rates and hotspots across the human genome. Science 310, 321–324 (2005).
Gabriel, S.B. et al. The structure of haplotype blocks in the human genome. Science 296, 2225–2229 (2002).
Ward, L.D. & Kellis, M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 40, D930–D934 (2012).
Boyle, A.P. et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 22, 1790–1797 (2012).
Cooper, G.M. et al. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 15, 901–913 (2005).
Yang, T.P. et al. Genevar: a database and Java application for the analysis and visualization of SNP-gene associations in eQTL studies. Bioinformatics 26, 2474–2476 (2010).
Acknowledgements
Leukaemia and Lymphoma Research provided principal funding for the study (LRF05001 and LRF06002). We acknowledge support from Cancer Research UK (C1298/A8362 supported by the Bobby Moore Fund) and the Arbib Fund. We also acknowledge National Health Service funding to the Royal Marsden/Institute of Cancer Research; National Institute for Health Research Biomedical Research Centre. The study made use of genotyping data on the 1958 Birth Cohort; a full list of the investigators who contributed to the generation of these data is available at http://www.wtccc.org.uk/. We thank L. Padyukov (Karolinska Institutet) and the Epidemiological Investigation of Rheumatoid Arthritis (EIRA) group for providing control samples from the Swedish population for the Swedish replication study. We are grateful to all clinicians for their involvement in patient ascertainment. This study makes use of data generated by the Wellcome Trust Case-Control Consortium 1 and 2. A full list of the investigators who contributed to the generation of the data is available at http://www.wtccc.org.uk/. We are grateful to all investigators and all the patients and individuals for their participation. We also thank the clinicians, other hospital staff and study staff that contributed to the blood sample and data collection for this study.
Author information
Authors and Affiliations
Contributions
R.S.H. obtained financial support, designed the project and provided overall project management. R.S.H. drafted the manuscript. H.E.S. performed project management and supervised genotyping. M.C.D.B. performed bioinformatic and statistical analyses. G.P.S. and A.H. performed genotyping. Y.W. and M.C.D.B. performed imputation analysis. D.C. and R.S.H. established the International CLL Linkage Consortium (ICLLLC). C.D. and D.C. performed recruitment of samples. In Sweden, L.M. performed sample collection and prepared DNA; R.R. performed collection of all cases and G.J. and K.E.S. performed sample collection in the Scandinavian Lymphoma Etiology (SCALE) study; and G.R. performed telomere analysis. In Newcastle, J.M.A. and D.J.A. conceived of the NCLLC; J.M.A. obtained financial support, supervised laboratory management and oversaw genotyping of cases with NCLLC; N.J.S. performed sample management of cases; A.G.H. developed the Newcastle Haematology Biobank, incorporating NCLLC; and T.M.-F., G.H.J., G.S., R.J.H., A.R.P., D.J.A., J.R.B., G.P., C.P. and C.F. developed protocols for recruitment of individuals with CLL and sample acquisition and performed sample collection of cases. In Leicester, M.J.S.D. performed overall management, collection and processing of samples; and S.J. and A.M. performed DNA extractions and IGVH mutation assays. All authors contributed to the final paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–7 and Supplementary Figures 1–5 (PDF 2934 kb)
Source data
Rights and permissions
About this article
Cite this article
Speedy, H., Di Bernardo, M., Sava, G. et al. A genome-wide association study identifies multiple susceptibility loci for chronic lymphocytic leukemia. Nat Genet 46, 56–60 (2014). https://doi.org/10.1038/ng.2843
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2843
This article is cited by
-
PASTRY: achieving balanced power for detecting risk and protective minor alleles in meta-analysis of association studies with overlapping subjects
BMC Bioinformatics (2024)
-
Multi-omics analysis identifies RFX7 targets involved in tumor suppression and neuronal processes
Cell Death Discovery (2023)
-
Implementation of individualised polygenic risk score analysis: a test case of a family of four
BMC Medical Genomics (2022)
-
Subclonal heterogeneity sheds light on the transformation trajectory in IGLV3-21R110 chronic lymphocytic leukemia
Blood Cancer Journal (2022)
-
TERT Gene rs2736100 and rs2736098 Polymorphisms are Associated with Increased Cancer Risk: A Meta-Analysis
Biochemical Genetics (2022)