Abstract
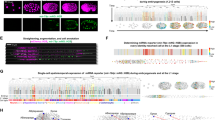
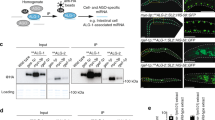
The complexity of multicellular organisms requires precise spatiotemporal regulation of gene expression during development. We find that in the nematode Caenorhabditis elegans approximately 2,000 transcripts undergo expression oscillations synchronized with larval transitions while thousands of genes are expressed in temporal gradients, similar to known timing regulators. By counting transcripts in individual worms, we show that pulsatile expression of the microRNA (miRNA) lin-4 maintains the temporal gradient of its target lin-14 by dampening its expression oscillations. Our results demonstrate that this insulation is optimal when pulsatile expression of the miRNA and its target is synchronous. We propose that such a miRNA-mediated incoherent feed-forward loop is a potent filter that prevents the propagation of potentially deleterious fluctuations in gene expression during the development of an organism.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Accession codes
References
Raj, A. & van Oudenaarden, A. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell 135, 216–226 (2008).
Rougvie, A.E. Intrinsic and extrinsic regulators of developmental timing: from miRNAs to nutritional cues. Development 132, 3787–3798 (2005).
Ruvkun, G. & Giusto, J. The Caenorhabditis elegans heterochronic gene lin-14 encodes a nuclear protein that forms a temporal developmental switch. Nature 338, 313–319 (1989).
Johnstone, I.L. & Barry, J.D. Temporal reiteration of a precise gene expression pattern during nematode development. EMBO J. 15, 3633–3639 (1996).
Gissendanner, C.R., Crossgrove, K., Kraus, K.A., Maina, C.V. & Sluder, A.E. Expression and function of conserved nuclear receptor genes in Caenorhabditis elegans. Dev. Biol. 266, 399–416 (2004).
Raizen, D.M. et al. Lethargus is a Caenorhabditis elegans sleep-like state. Nature 451, 569–572 (2008).
Page, A.P. & Johnstone, I.L. The cuticle. WormBook http://www.wormbook.org/ (2007). 10.1895/wormbook.1.138.1.
Altun, Z.F. & Hall, D.H. Introduction. WormAtlas http://www.wormatlas.org (2009).
Hubbard, E.J.A. & Grenstein, D. Introduction to the germ line. WormBook http://www.wormbook.org/ (2005). 10.1895/wormbook.1.18.1.
Ambros, V. & Horvitz, H.R. The lin-14 locus of Caenorhabditis elegans controls the time of expression of specific postembryonic developmental events. Genes Dev. 1, 398–414 (1987).
Moss, E.G., Lee, R.C. & Ambros, V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell 88, 637–646 (1997).
Slack, F.J. et al. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol. Cell 5, 659–669 (2000).
Ambros, V. & Horvitz, H. Heterochronic mutants of the nematode Caenorhabditis elegans. Science 226, 409–416 (1984).
Ambros, V. A hierarchy of regulatory genes controls a larva-to-adult developmental switch in C. elegans. Cell 57, 49–57 (1989).
Lee, R.C., Feinbaum, R.L. & Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854 (1993).
Bartel, D.P. MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 (2009).
Reinhart, B.J. & Ruvkun, G. Isoform-specific mutations in the Caenorhabditis elegans heterochronic gene lin-14 affect stage-specific patterning. Genetics 157, 199–209 (2001).
Alon, U. Network motifs: theory and experimental approaches. Nat. Rev. Genet. 8, 450–461 (2007).
Gerstein, M.B. et al. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science 330, 1775–1787 (2010).
Mortazavi, A., Williams, B.A., Mccue, K., Schaeffer, L. & Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 1–8 (2008).
Sönnichsen, B. et al. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature 434, 462–469 (2005).
Lewis, B.P., Burge, C.B. & Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20 (2005).
Selbach, M. et al. Widespread changes in protein synthesis induced by microRNAs. Nature 455, 58–63 (2008).
Baek, D. et al. The impact of microRNAs on protein output. Nature 455, 64–71 (2008).
Bagga, S. et al. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 122, 553–563 (2005).
Lim, L.P. et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433, 769–773 (2005).
Dreszer, T.R. et al. The UCSC Genome Browser database: extensions and updates 2011. Nucleic Acids Res. 40, D918–D923 (2012).
Zhang, X., Zabinsky, R., Teng, Y., Cui, M. & Han, M. microRNAs play critical roles in the survival and recovery of Caenorhabditis elegans from starvation-induced L1 diapause. Proc. Natl. Acad. Sci. USA 108, 17997–18002 (2011).
de Lencastre, A. et al. MicroRNAs both promote and antagonize longevity in C. elegans. Curr. Biol. 20, 2159–2168 (2010).
Boulias, K. & Horvitz, H.R. The C. elegans microRNA mir-71 acts in neurons to promote germline-mediated longevity through regulation of DAF-16/FOXO. Cell Metab. 15, 439–450 (2012).
Alvarez-Saavedra, E. & Horvitz, H.R. Many families of C. elegans microRNAs are not essential for development or viability. Curr. Biol. 20, 367–373 (2010).
Martinez, N.J. et al. Genome-scale spatiotemporal analysis of Caenorhabditis elegans microRNA promoter activity. Genome Res. 18, 2005–2015 (2008).
Feinbaum, R. & Ambros, V. The timing of lin-4 RNA accumulation controls the timing of postembryonic developmental events in Caenorhabditis elegans. Dev. Biol. 210, 87–95 (1999).
Raj, A., Bogaard, P.V.D., Rifkin, S.A., Oudenaarden, A.V. & Tyagi, S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods 5, 877–879 (2008).
Knight, C.G., Patel, M.N., Azevedo, R.B.R. & Leroi, A.M. A novel mode of ecdysozoan growth in Caenorhabditis elegans. Evol. Dev. 4, 16–27 (2002).
Sulston, J.E. & Horvitz, H.R. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56, 110–156 (1977).
Ruvkun, G. et al. Molecular genetics of the Caenorhabditis elegans heterochronic gene lin-14. Genetics 121, 501–516 (1989).
Wightman, B., Ha, I. & Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75, 855–862 (1993).
Zhang, H. & Fire, A.Z. Cell autonomous specification of temporal identity by Caenorhabditis elegans microRNA lin-4. Dev. Biol. 344, 603–610 (2010).
Jeon, M. Similarity of the C. elegans developmental timing protein LIN-42 to circadian rhythm proteins. Science 286, 1141–1146 (1999).
Frand, A.R., Russel, S. & Ruvkun, G. Functional genomic analysis of C. elegans molting. PLoS Biol. 3, e312 (2005).
Basu, S., Mehreja, R., Thiberge, S., Chen, M.-T. & Weiss, R. Spatiotemporal control of gene expression with pulse-generating networks. Proc. Natl. Acad. Sci. USA 101, 6355–6360 (2004).
Tsang, J., Zhu, J. & van Oudenaarden, A. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol. Cell 26, 753–767 (2007).
Li, H. & Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010).
Kohonen, T. Self-organized formation of topologically correct feature maps. Biol. Cybern. 43, 59–69 (1982).
Altschul, S.F., Gish, W., Miller, W., Myers, E.W. & Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Falcon, S. & Gentleman, R. Using GOstats to test gene lists for GO term association. Bioinformatics 23, 257–258 (2007).
Acknowledgements
We thank V. Ambros, H.R. Horvitz, G. Ruvkun and J. Gore for helpful discussions and advice. We thank S. Itzkovitz, J.P. Junker, G. Neuert, J. van Zon, A. Sahay and C. Engert for a critical reading of the manuscript. We thank the Massachusetts Institute of Technology (MIT) BioMicro Center for Illumina sample preparation and sequencing experiments. We also thank the Biopolymers & Proteomics Core Facility of the MIT Koch Institute for Integrative Cancer Research for probe purification. Several nematode strains used in this work were provided by H.R. Horvitz (Massachusetts Institute of Technology) and A. Fire (Stanford University) and the Caenorhabditis Genetics Center, which is funded by the US National Institutes of Health (NIH) and National Center for Research Resources (NCRR). This work was supported by the US NIH/National Cancer Institute Physical Sciences Oncology Center at MIT (U54CA143874), a US NIH Pioneer award (8 DP1 CA174420-05), a European Research Council Advanced grant (ERC-AdG 294325-GeneNoiseControl) and a Green Cross Corp. (Republic of Korea) Mogam Science Scholarship.
Author information
Authors and Affiliations
Contributions
D.h.K. and A.v.O. conceived the project and designed the experiments. D.h.K. performed the experiments. D.h.K., D.G. and A.v.O. analyzed the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Note and Supplementary Figures 1–14 (PDF 6048 kb)
Supplementary Table 1
Assignment of genes to expression clusters for clusters genes (XLSX 271 kb)
Supplementary Table 2
A full list of GO terms enriched in clusters of coexpressed genes (XLSX 300 kb)
Rights and permissions
About this article
Cite this article
Kim, D., Grün, D. & van Oudenaarden, A. Dampening of expression oscillations by synchronous regulation of a microRNA and its target. Nat Genet 45, 1337–1344 (2013). https://doi.org/10.1038/ng.2763
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2763
This article is cited by
-
Deviations from temporal scaling support a stage-specific regulation for C. elegans postembryonic development
BMC Biology (2022)
-
Coupling of growth rate and developmental tempo reduces body size heterogeneity in C. elegans
Nature Communications (2022)
-
Real age prediction from the transcriptome with RAPToR
Nature Methods (2022)
-
Integrated microRNA and mRNA analysis in the pathogenic filamentous fungus Trichophyton rubrum
BMC Genomics (2018)
-
Contribution of trans regulatory eQTL to cryptic genetic variation in C. elegans
BMC Genomics (2017)