Abstract
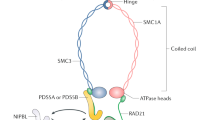
Cohesin is a multimeric protein complex that is involved in the cohesion of sister chromatids, post-replicative DNA repair and transcriptional regulation. Here we report recurrent mutations and deletions involving multiple components of the cohesin complex, including STAG2, RAD21, SMC1A and SMC3, in different myeloid neoplasms. These mutations and deletions were mostly mutually exclusive and occurred in 12.1% (19/157) of acute myeloid leukemia, 8.0% (18/224) of myelodysplastic syndromes, 10.2% (9/88) of chronic myelomonocytic leukemia, 6.3% (4/64) of chronic myelogenous leukemia and 1.3% (1/77) of classical myeloproliferative neoplasms. Cohesin-mutated leukemic cells showed reduced amounts of chromatin-bound cohesin components, suggesting a substantial loss of cohesin binding sites on chromatin. The growth of leukemic cell lines harboring a mutation in RAD21 (Kasumi-1 cells) or having severely reduced expression of RAD21 and STAG2 (MOLM-13 cells) was suppressed by forced expression of wild-type RAD21 and wild-type RAD21 and STAG2, respectively. These findings suggest a role for compromised cohesin functions in myeloid leukemogenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Bejar, R., Levine, R. & Ebert, B.L. Unraveling the molecular pathophysiology of myelodysplastic syndromes. J. Clin. Oncol. 29, 504–515 (2011).
Marcucci, G., Haferlach, T. & Dohner, H. Molecular genetics of adult acute myeloid leukemia: prognostic and therapeutic implications. J. Clin. Oncol. 29, 475–486 (2011).
Yoshida, K. et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 478, 64–69 (2011).
Gruber, S., Haering, C.H. & Nasmyth, K. Chromosomal cohesin forms a ring. Cell 112, 765–777 (2003).
Nasmyth, K. & Haering, C.H. Cohesin: its roles and mechanisms. Annu. Rev. Genet. 43, 525–558 (2009).
Ström, L. et al. Postreplicative formation of cohesion is required for repair and induced by a single DNA break. Science 317, 242–245 (2007).
Watrin, E. & Peters, J.M. The cohesin complex is required for the DNA damage-induced G2/M checkpoint in mammalian cells. EMBO J. 28, 2625–2635 (2009).
Dorsett, D. Cohesin, gene expression and development: lessons from Drosophila. Chromosome Res. 17, 185–200 (2009).
Dorsett, D. et al. Effects of sister chromatid cohesion proteins on cut gene expression during wing development in Drosophila. Development 132, 4743–4753 (2005).
Horsfield, J.A. et al. Cohesin-dependent regulation of Runx genes. Development 134, 2639–2649 (2007).
Parelho, V. et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell 132, 422–433 (2008).
Wendt, K.S. et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 451, 796–801 (2008).
Bose, T. & Gerton, J.L. Cohesinopathies, gene expression, and chromatin organization. J. Cell Biol. 189, 201–210 (2010).
Deardorff, M.A. et al. HDAC8 mutations in Cornelia de Lange syndrome affect the cohesin acetylation cycle. Nature 489, 313–317 (2012).
Deardorff, M.A. et al. RAD21 mutations cause a human cohesinopathy. Am. J. Hum. Genet. 90, 1014–1027 (2012).
Solomon, D.A. et al. Mutational inactivation of STAG2 causes aneuploidy in human cancer. Science 333, 1039–1043 (2011).
Beckouët, F. et al. An Smc3 acetylation cycle is essential for establishment of sister chromatid cohesion. Mol. Cell 39, 689–699 (2010).
Liu, J. et al. Transcriptional dysregulation in NIPBL and cohesin mutant human cells. PLoS Biol. 7, e1000119 (2009).
Liu, J. et al. Genome-wide DNA methylation analysis in cohesin mutant human cell lines. Nucleic Acids Res. 38, 5657–5671 (2010).
Schaaf, C.A. et al. Regulation of the Drosophila enhancer of split and invected-engrailed gene complexes by sister chromatid cohesion proteins. PLoS ONE 4, e6202 (2009).
Ding, L. et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 481, 506–510 (2012).
Walter, M.J. et al. Clonal architecture of secondary acute myeloid leukemia. N. Engl. J. Med. 366, 1090–1098 (2012).
Welch, J.S. et al. The origin and evolution of mutations in acute myeloid leukemia. Cell 150, 264–278 (2012).
The Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 368, 2059–2074 (2013).
Walter, M.J. et al. Clonal diversity of recurrently mutated genes in myelodysplastic syndromes. Leukemia 27, 12785–1282 (2013).
Barber, T.D. et al. Chromatid cohesion defects may underlie chromosome instability in human colorectal cancers. Proc. Natl. Acad. Sci. USA 105, 3443–3448 (2008).
Heidinger-Pauli, J.M., Mert, O., Davenport, C., Guacci, V. & Koshland, D. Systematic reduction of cohesin differentially affects chromosome segregation, condensation, and DNA repair. Curr. Biol. 20, 957–963 (2010).
Hadjur, S. et al. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature 460, 410–413 (2009).
Chan, D.A. & Giaccia, A.J. Harnessing synthetic lethal interactions in anticancer drug discovery. Nat. Rev. Drug Discov. 10, 351–364 (2011).
Sato, Y. et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat. Genet. doi:10.1038/ng.2699 (24 June 2013).
Kent, W.J. BLAT—the BLAST-like alignment tool. Genome Res. 12, 656–664 (2002).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Kumar, P., Henikoff, S. & Ng, P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 4, 1073–1081 (2009).
Adzhubei, I.A. et al. A method and server for predicting damaging missense mutations. Nat. Methods 7, 248–249 (2010).
Schwarz, J.M., Rodelsperger, C., Schuelke, M. & Seelow, D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods 7, 575–576 (2010).
Ronaghi, M. Pyrosequencing sheds light on DNA sequencing. Genome Res. 11, 3–11 (2001).
Shih, L.Y. et al. Emerging kinetics of BCR-ABL1 mutations and their effect on disease outcomes in chronic myeloid leukemia patients with imatinib failure. Leuk. Res. 37, 43–49 (2013).
Qin, J., Jones, R.C. & Ramakrishnan, R. Studying copy number variations using a nanofluidic platform. Nucleic Acids Res. 36, e116 (2008).
Dube, S., Qin, J. & Ramakrishnan, R. Mathematical analysis of copy number variation in a DNA sample using digital PCR on a nanofluidic device. PLoS ONE 3, e2876 (2008).
Totoki, Y. et al. High-resolution characterization of a hepatocellular carcinoma genome. Nat. Genet. 43, 464–469 (2011).
Nannya, Y. et al. A robust algorithm for copy number detection using high-density oligonucleotide single nucleotide polymorphism genotyping arrays. Cancer Res. 65, 6071–6079 (2005).
Yamamoto, G. et al. Highly sensitive method for genomewide detection of allelic composition in nonpaired, primary tumor specimens by use of affymetrix single-nucleotide-polymorphism genotyping microarrays. Am. J. Hum. Genet. 81, 114–126 (2007).
Hosoya, N. et al. Genomewide screening of DNA copy number changes in chronic myelogenous leukemia with the use of high-resolution array-based comparative genomic hybridization. Genes Chromosom. Cancer 45, 482–494 (2006).
Sanada, M. et al. Gain-of-function of mutated C-CBL tumour suppressor in myeloid neoplasms. Nature 460, 904–908 (2009).
Nagae, G. et al. Tissue-specific demethylation in CpG-poor promoters during cellular differentiation. Hum. Mol. Genet. 20, 2710–2721 (2011).
Nabekura, T., Otsu, M., Nagasawa, T., Nakauchi, H. & Onodera, M. Potent vaccine therapy with dendritic cells genetically modified by the gene-silencing–resistant retroviral vector GCDNsap. Mol. Ther. 13, 301–309 (2006).
Agarwal, S. et al. Isolation, characterization, and genetic complementation of a cellular mutant resistant to retroviral infection. Proc. Natl. Acad. Sci. USA 103, 15933–15938 (2006).
Li, C. & Wong, W.H. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl. Acad. Sci. USA 98, 31–36 (2001).
Mortazavi, A., Williams, B.A., McCue, K., Schaeffer, L. & Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628 (2008).
Pruitt, K.D., Tatusova, T., Brown, G.R. & Maglott, D.R. NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 40, D130–D135 (2012).
Acknowledgements
This work was supported by Grants-in-Aid from the Ministry of Health, Labor and Welfare of Japan and KAKENHI (23249052, 22134006 and 21790907; S.O.), the Industrial Technology Research Grant Program from the New Energy and Industrial Technology Development Organization (NEDO; S.O.) (08C46598a), NHRI-EX100-10003NI Taiwan (L.-Y.S.), the project for development of innovative research on cancer therapies (p-direct; S.O.) and the Japan Society for the Promotion of Science through the Funding Program for World-Leading Innovative R&D on Science and Technology, initiated by the Council for Science and Technology Policy (CSTP; S.O.). We thank Y. Hayashi (Gunma Children's Medical Centre), R.C. Mulligan (Harvard Medical School), S. Sugano (The University of Tokyo), M. Onodera (National Center for Child Health and Development, Japan) and L. Ström (Karolinska Institute) for providing materials. We thank Y. Yamazaki for cell sorting. We also thank Y. Mori, M. Nakamura, N. Mizota and S. Ichimura for their technical assistance and M. Ueda for encouragement.
Author information
Authors and Affiliations
Contributions
A.K., Y.N., K.Y., A.S.-O., Y. Sato and M.S. processed and analyzed genetic materials and performed sequencing and SNP array analysis. Y. Shiraishi, Y.O., R.N., A.S.-O., H.T., T.S., K.C., M.N. and S. Miyano performed bioinformatics analyses of the sequencing data. L.-Y.S. performed pyrosequencing analysis, and A.N. and S.I. performed digital PCR. G.N. and H.A. performed methylation analysis. M.M., M.B. and K.S. performed studies on protein expression of cohesin components. A.K., M.S., T.Y., R.Y., M.O. and H.N. were involved in the functional studies. A.K. and A.S.-O. performed expression microarray experiments and their analyses. L.-Y.S., D.N., T.A., C.H., F.N., W.-K.H., T.H., H.P.K., T.N., H.M., S. Miyawaki, M.S.-Y., K.I., N.O. and S.C. collected specimens and were involved in project planning. A.K., L.-Y.S., M.M., A.S.-O. and S.O. generated figures and tables. S.O. led the entire project, and A.K. and S.O. wrote the manuscript. All authors participated in the discussion and interpretation of the data.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–13 and Supplementary Tables 1–18 (PDF 3333 kb)
Source data
Rights and permissions
About this article
Cite this article
Kon, A., Shih, LY., Minamino, M. et al. Recurrent mutations in multiple components of the cohesin complex in myeloid neoplasms. Nat Genet 45, 1232–1237 (2013). https://doi.org/10.1038/ng.2731
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2731
This article is cited by
-
The synergism of SMC1A cohesin gene silencing and bevacizumab against colorectal cancer
Journal of Experimental & Clinical Cancer Research (2024)
-
Long-range gene regulation in hormone-dependent cancer
Nature Reviews Cancer (2023)
-
Context-dependent perturbations in chromatin folding and the transcriptome by cohesin and related factors
Nature Communications (2023)
-
Cohesin maintains replication timing to suppress DNA damage on cancer genes
Nature Genetics (2023)
-
Epigenetic regulation by ASXL1 in myeloid malignancies
International Journal of Hematology (2023)