Abstract
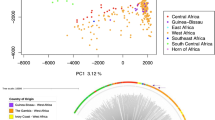
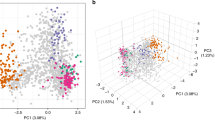
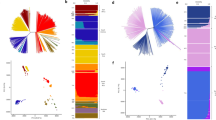
We describe an analysis of genome variation in 825 P. falciparum samples from Asia and Africa that identifies an unusual pattern of parasite population structure at the epicenter of artemisinin resistance in western Cambodia. Within this relatively small geographic area, we have discovered several distinct but apparently sympatric parasite subpopulations with extremely high levels of genetic differentiation. Of particular interest are three subpopulations, all associated with clinical resistance to artemisinin, which have skewed allele frequency spectra and high levels of haplotype homozygosity, indicative of founder effects and recent population expansion. We provide a catalog of SNPs that show high levels of differentiation in the artemisinin-resistant subpopulations, including codon variants in transporter proteins and DNA mismatch repair proteins. These data provide a population-level genetic framework for investigating the biological origins of artemisinin resistance and for defining molecular markers to assist in its elimination.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
White, N.J. Artemisinin resistance—the clock is ticking. Lancet 376, 2051–2052 (2010).
Payne, D. Spread of chloroquine resistance in Plasmodium falciparum. Parasitol. Today 3, 241–246 (1987).
Roper, C. et al. Intercontinental spread of pyrimethamine-resistant malaria. Science 305, 1124 (2004).
Mita, T. et al. Limited geographical origin and global spread of sulfadoxine-resistant dhps alleles in Plasmodium falciparum populations. J. Infect. Dis. 204, 1980–1988 (2011).
Noedl, H. et al. Evidence of artemisinin-resistant malaria in western Cambodia. N. Engl. J. Med. 359, 2619–2620 (2008).
Dondorp, A.M. et al. Artemisinin resistance in Plasmodium falciparum malaria. N. Engl. J. Med. 361, 455–467 (2009).
Cheeseman, I.H. et al. A major genome region underlying artemisinin resistance in malaria. Science 336, 79–82 (2012).
Amaratunga, C. et al. Artemisinin-resistant Plasmodium falciparum in Pursat province, western Cambodia: a parasite clearance rate study. Lancet Infect. Dis. 12, 851–858 (2012).
Phyo, A.P. et al. Emergence of artemisinin-resistant malaria on the western border of Thailand: a longitudinal study. Lancet 379, 1960–1966 (2012).
Tran, T.H. et al. In vivo susceptibility of Plasmodium falciparum to artesunate in Binh Phuoc Province, Vietnam. Malar. J. 11, 355 (2012).
Anderson, T.J. et al. High heritability of malaria parasite clearance rate indicates a genetic basis for artemisinin resistance in western Cambodia. J. Infect. Dis. 201, 1326–1330 (2010).
Dondorp, A.M. et al. The threat of artemisinin-resistant malaria. N. Engl. J. Med. 365, 1073–1075 (2011).
Manske, M. et al. Analysis of Plasmodium falciparum diversity in natural infections by deep sequencing. Nature 487, 375–379 (2012).
Hay, S.I. et al. A world malaria map: Plasmodium falciparum endemicity in 2007. PLoS Med. 6, e1000048 (2009).
Flegg, J.A., Guerin, P.J., White, N.J. & Stepniewska, K. Standardizing the measurement of parasite clearance in falciparum malaria: the parasite clearance estimator. Malar. J. 10, 339 (2011).
Mu, J. et al. Multiple transporters associated with malaria parasite responses to chloroquine and quinine. Mol. Microbiol. 49, 977–989 (2003).
Mu, J. et al. Plasmodium falciparum genome-wide scans for positive selection, recombination hot spots and resistance to antimalarial drugs. Nat. Genet. 42, 268–271 (2010).
Anderson, T.J. et al. Are transporter genes other than the chloroquine resistance locus (pfcrt) and multidrug resistance gene (pfmdr) associated with antimalarial drug resistance? Antimicrob. Agents Chemother. 49, 2180–2188 (2005).
Fidock, D.A. et al. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol. Cell 6, 861–871 (2000).
Lim, P. et al. pfcrt polymorphism and chloroquine resistance in Plasmodium falciparum strains isolated in Cambodia. Antimicrob. Agents Chemother. 47, 87–94 (2003).
Isozumi, R. et al. Longitudinal survey of Plasmodium falciparum infection in Vietnam: characteristics of antimalarial resistance and their associated factors. J. Clin. Microbiol. 48, 70–77 (2010).
Foote, S.J. et al. Several alleles of the multidrug-resistance gene are closely linked to chloroquine resistance in Plasmodium falciparum. Nature 345, 255–258 (1990).
Peel, S.A., Bright, P., Yount, B., Handy, J. & Baric, R.S. A strong association between mefloquine and halofantrine resistance and amplification, overexpression, and mutation in the P-glycoprotein gene homolog (pfmdr) of Plasmodium falciparum in vitro. Am. J. Trop. Med. Hyg. 51, 648–658 (1994).
Preechapornkul, P. et al. Plasmodium falciparum pfmdr1 amplification, mefloquine resistance, and parasite fitness. Antimicrob. Agents Chemother. 53, 1509–1515 (2009).
Plowe, C.V. et al. Mutations in Plasmodium falciparum dihydrofolate reductase and dihydropteroate synthase and epidemiologic patterns of pyrimethamine-sulfadoxine use and resistance. J. Infect. Dis. 176, 1590–1596 (1997).
Vinayak, S. et al. Origin and evolution of sulfadoxine resistant Plasmodium falciparum. PLoS Pathog. 6, e1000830 (2010).
Kublin, J.G. et al. Molecular assays for surveillance of antifolate-resistant malaria. Lancet 351, 1629–1630 (1998).
Triglia, T., Wang, P., Sims, P.F., Hyde, J.E. & Cowman, A.F. Allelic exchange at the endogenous genomic locus in Plasmodium falciparum proves the role of dihydropteroate synthase in sulfadoxine-resistant malaria. EMBO J. 17, 3807–3815 (1998).
Peterson, D.S., Walliker, D. & Wellems, T.E. Evidence that a point mutation in dihydrofolate reductase–thymidylate synthase confers resistance to pyrimethamine in falciparum malaria. Proc. Natl. Acad. Sci. USA 85, 9114–9118 (1988).
Plowe, C.V. The evolution of drug-resistant malaria. Trans. R. Soc. Trop. Med. Hyg. 103 (suppl. 1), S11–S14 (2009).
Rathod, P.K., McErlean, T. & Lee, P.C. Variations in frequencies of drug resistance in Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 94, 9389–9393 (1997).
Jiricny, J. Replication errors: cha(lle)nging the genome. EMBO J. 17, 6427–6436 (1998).
Shankar, J. & Tuteja, R. UvrD helicase of Plasmodium falciparum. Gene 410, 223–233 (2008).
Hall, M.C., Jordan, J.R. & Matson, S.W. Evidence for a physical interaction between the Escherichia coli methyl-directed mismatch repair proteins MutL and UvrD. EMBO J. 17, 1535–1541 (1998).
Mechanic, L.E., Frankel, B.A. & Matson, S.W. Escherichia coli MutL loads DNA helicase II onto DNA. J. Biol. Chem. 275, 38337–38346 (2000).
Anderson, T.J. et al. Microsatellite markers reveal a spectrum of population structures in the malaria parasite Plasmodium falciparum. Mol. Biol. Evol. 17, 1467–1482 (2000).
Dye, C. & Williams, B.G. Multigenic drug resistance among inbred malaria parasites. Proc. Biol. Sci. 264, 61–67 (1997).
Sinka, M.E. et al. The dominant Anopheles vectors of human malaria in the Asia-Pacific region: occurrence data, distribution maps and bionomic precis. Parasit. Vectors 4, 89 (2011).
Verdrager, J. Epidemiology of the emergence and spread of drug-resistant falciparum malaria in South-East Asia and Australasia. J. Trop. Med. Hyg. 89, 277–289 (1986).
Payne, D. Did medicated salt hasten the spread of chloroquine resistance in Plasmodium falciparum? Parasitol. Today 4, 112–115 (1988).
World Health Organization. Recommendations. in Global Plan for Artemisinin Resistance Containment 23–27 (World Health Organization, Geneva, 2011).
Bethell, D. et al. Artesunate dose escalation for the treatment of uncomplicated malaria in a region of reported artemisinin resistance: a randomized clinical trial. PLoS ONE 6, e19283 (2011).
Aurrecoechea, C. et al. PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res. 37, D539–D543 (2009).
Lawson, D.J., Hellenthal, G., Myers, S. & Falush, D. Inference of population structure using dense haplotype data. PLoS Genet. 8, e1002453 (2012).
Alexander, D.H., Novembre, J. & Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664 (2009).
Acknowledgements
We would like to thank V. Cornelius and R. Giacomantonio for their support in producing and reviewing this manuscript; S. Uk and E.S. Phelps for their contributions in the Cambodian studies; and T. Anderson for helpful review comments. The sequencing, genotyping and analysis components of this study were supported by the Wellcome Trust through core funding of the Wellcome Trust Sanger Institute (098051), core funding of the Wellcome Trust Centre for Human Genetics (090532/Z/09/Z) and a Strategic Award (090770/Z/09/Z) and the MRC through the MRC Centre for Genomics and Global Health (G0600718) and an MRC Professorship to D.P.K. (G19/9). Other parts of this study were partly supported by the Wellcome Trust; the MRC; the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, US National Institutes of Health; and a Howard Hughes Medical Institute International Scholarship (55005502) to A.A.D. P.R. is a staff member of the World Health Organization; he alone is responsible for the views expressed in this publication, and they do not necessarily represent the decisions, policy or views of the World Health Organization.
Author information
Authors and Affiliations
Contributions
S.C., C.A., P.L., S. Suon, S. Sreng, J.M.A., S.D., C.N., C.M.C., D.S., Y.S., C.L., M.M.F., L.A.-E., A.V.O.H., V.A., M.I., F.N., X.S., P.R., F.A., C.D., T.T.H., M.F.B., C.Q.T., A.A.-N., D.J.C., A.A.D., O.K.D., I.Z., J.-B.O., S.A., N.P.D., N.J.W., D.B., A.M.D., C.V.P. and R.M.F. carried out field and laboratory studies to obtain P. falciparum samples for sequencing. C.A., P.L., S. Suon, S. Sreng, J.M.A., S.D., C.N., C.M.C., D.S., Y.S., C.L., M.M.F., F.N., X.S., P.R., F.A., N.J.W., D.B., A.M.D., C.V.P. and R.M.F. carried out clinical studies to obtain ART phenotype data. S.C., D.A., E.D., M.S., S.A., O.K., S.O.O., B.M., C.I.N. and M.B. developed and implemented methods for sample processing and sequencing library preparation. O.M., J.A.-G., M.M., G. Maslen, V.R.-R., D.J. and A.M. developed software for data management and visualization. K.A.R., C.H., D.A. and M.M. carried out validation experiments. C.V.P., S.T.-H., G. McVean and R.M.F. contributed to development of the project. B.M., M.B., C.I.N. and J.C.R. provided project management and oversight. O.M., J.A.-G., M.M., J.O., C.G. and C.C.A.S. carried out data analyses. D.P.K., O.M. and J.A.-G. wrote the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–9, Supplementary Tables 1–6 and 8–14, Supplementary Note (PDF 1595 kb)
Supplementary Table 7
Lists of SNPs that are highly differentiated in the Cambodian outlier subpopulations, compared to KH1. (XLSX 137 kb)
Supplementary Table 8
Lists of SNP that are highly differentiated between the Cambodian subpopulations. (XLSX 210 kb)
Rights and permissions
About this article
Cite this article
Miotto, O., Almagro-Garcia, J., Manske, M. et al. Multiple populations of artemisinin-resistant Plasmodium falciparum in Cambodia. Nat Genet 45, 648–655 (2013). https://doi.org/10.1038/ng.2624
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2624
This article is cited by
-
In vitro delayed response to dihydroartemisinin of malaria parasites infecting sickle cell erythocytes
Malaria Journal (2024)
-
Genomics reveals heterogeneous Plasmodium falciparum transmission and selection signals in Zambia
Communications Medicine (2024)
-
MalariaSED: a deep learning framework to decipher the regulatory contributions of noncoding variants in malaria parasites
Genome Biology (2023)
-
Artemisinin derivatives induce oxidative stress leading to DNA damage and caspase-mediated apoptosis in Theileria annulata-transformed cells
Cell Communication and Signaling (2023)
-
Genomics of Plasmodium vivax in Colombia reveals evidence of local bottle-necking and inter-country connectivity in the Americas
Scientific Reports (2023)