Abstract
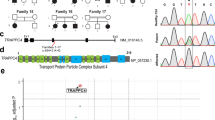
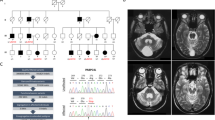
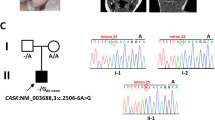
Static encephalopathy of childhood with neurodegeneration in adulthood (SENDA) is a recently established subtype of neurodegeneration with brain iron accumulation (NBIA)1,2,3. By exome sequencing, we found de novo heterozygous mutations in WDR45 at Xp11.23 in two individuals with SENDA, and three additional WDR45 mutations were identified in three other subjects by Sanger sequencing. Using lymphoblastoid cell lines (LCLs) derived from the subjects, aberrant splicing was confirmed in two, and protein expression was observed to be severely impaired in all five. WDR45 encodes WD-repeat domain 45 (WDR45). WDR45 (also known as WIPI4) is one of the four mammalian homologs of yeast Atg18, which has an important role in autophagy4,5. Lower autophagic activity and accumulation of aberrant early autophagic structures were demonstrated in the LCLs of the affected subjects. These findings provide direct evidence that an autophagy defect is indeed associated with a neurodegenerative disorder in humans.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
References
Gregory, A., Polster, B.J. & Hayflick, S.J. Clinical and genetic delineation of neurodegeneration with brain iron accumulation. J. Med. Genet. 46, 73–80 (2009).
Kruer, M.C. et al. Neuroimaging features of neurodegeneration with brain iron accumulation. AJNR Am. J. Neuroradiol. 33, 407–414 (2012).
Schneider, S.A. & Bhatia, K.P. Syndromes of neurodegeneration with brain iron accumulation. Semin. Pediatr. Neurol. 19, 57–66 (2012).
Polson, H.E. et al. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy 6, 506–522 (2010).
Lu, Q. et al. The WD40 repeat PtdIns(3)P-binding protein EPG-6 regulates progression of omegasomes to autophagosomes. Dev. Cell 21, 343–357 (2011).
Gregory, A. & Hayflick, S.J. Genetics of neurodegeneration with brain iron accumulation. Curr. Neurol. Neurosci. Rep. 11, 254–261 (2011).
Kimura, Y. et al. MRI, MR spectroscopy, and diffusion tensor imaging findings in patient with static encephalopathy of childhood with neurodegeneration in adulthood (SENDA). Brain Dev. published online; doi:10.1016/j.braindev.2012.07.008 (11 August 2012).
Kasai-Yoshida, E. et al. First video report of static encephalopathy of childhood with neurodegeneration in adulthood. Mov. Disord. published online; doi:10.1002/mds.25158 (6 February 2013).
Mizushima, N. & Komatsu, M. Autophagy: renovation of cells and tissues. Cell 147, 728–741 (2011).
Nakatogawa, H., Suzuki, K., Kamada, Y. & Ohsumi, Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat. Rev. Mol. Cell Biol. 10, 458–467 (2009).
Xie, Z. & Klionsky, D.J. Autophagosome formation: core machinery and adaptations. Nat. Cell Biol. 9, 1102–1109 (2007).
Mizushima, N., Yoshimori, T. & Ohsumi, Y. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27, 107–132 (2011).
Baskaran, S., Ragusa, M.J., Boura, E. & Hurley, J.H. Two-site recognition of phosphatidylinositol 3-phosphate by PROPPINs in autophagy. Mol. Cell 47, 339–348 (2012).
Krick, R. et al. Structural and functional characterization of the two phosphoinositide binding sites of PROPPINs, a β-propeller protein family. Proc. Natl. Acad. Sci. USA 109, E2042–E2049 (2012).
Watanabe, Y. et al. Structure-based analyses reveal distinct binding sites for Atg2 and phosphoinositides in Atg18. J. Biol. Chem. 287, 31681–31690 (2012).
Suzuki, K., Kubota, Y., Sekito, T. & Ohsumi, Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells 12, 209–218 (2007).
Velikkakath, A.K., Nishimura, T., Oita, E., Ishihara, N. & Mizushima, N. Mammalian Atg2 proteins are essential for autophagosome formation and important for regulation of size and distribution of lipid droplets. Mol. Biol. Cell 23, 896–909 (2012).
Mizushima, N., Yoshimori, T. & Levine, B. Methods in mammalian autophagy research. Cell 140, 313–326 (2010).
Itakura, E., Kishi-Itakura, C., Koyama-Honda, I. & Mizushima, N. Structures containing Atg9A and the ULK1 complex independently target depolarized mitochondria at initial stages of Parkin-mediated mitophagy. J. Cell Sci. 125, 1488–1499 (2012).
Orsi, A. et al. Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy. Mol. Biol. Cell 23, 1860–1873 (2012).
Hara, T. et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441, 885–889 (2006).
Komatsu, M. et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441, 880–884 (2006).
Menzies, F.M., Moreau, K. & Rubinsztein, D.C. Protein misfolding disorders and macroautophagy. Curr. Opin. Cell Biol. 23, 190–197 (2011).
Valente, E.M. et al. Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science 304, 1158–1160 (2004).
Kitada, T. et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 392, 605–608 (1998).
Youle, R.J. & van der Bliek, A.M. Mitochondrial fission, fusion, and stress. Science 337, 1062–1065 (2012).
Youle, R.J. & Narendra, D.P. Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 12, 9–14 (2011).
Kuma, A. et al. The role of autophagy during the early neonatal starvation period. Nature 432, 1032–1036 (2004).
Saitoh, T. et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1β production. Nature 456, 264–268 (2008).
Saitoh, T. et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc. Natl. Acad. Sci. USA 106, 20842–20846 (2009).
Sou, Y.S. et al. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol. Biol. Cell 19, 4762–4775 (2008).
Komatsu, M. et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 169, 425–434 (2005).
Haack, T.B. et al. Exome sequencing reveals de novo WDR45 mutations causing a phenotypically distinct, X-linked dominant form of NBIA. Am. J. Hum. Genet. 91, 1144–1149 (2012).
Proikas-Cezanne, T. et al. WIPI-1α (WIPI49), a member of the novel 7-bladed WIPI protein family, is aberrantly expressed in human cancer and is linked to starvation-induced autophagy. Oncogene 23, 9314–9325 (2004).
DePristo, M.A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010).
Saitsu, H. et al. STXBP1 mutations in early infantile epileptic encephalopathy with suppression-burst pattern. Epilepsia 51, 2397–2405 (2010).
Kondo, Y. et al. A family of oculofaciocardiodental syndrome (OFCD) with a novel BCOR mutation and genomic rearrangements involving NHS. J. Hum. Genet. 57, 197–201 (2012).
Allen, R.C., Zoghbi, H.Y., Moseley, A.B., Rosenblatt, H.M. & Belmont, J.W. Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human androgen-receptor gene correlates with X chromosome inactivation. Am. J. Hum. Genet. 51, 1229–1239 (1992).
Carrel, L. & Willard, H.F. An assay for X inactivation based on differential methylation at the fragile X locus, FMR1. Am. J. Med. Genet. 64, 27–30 (1996).
Hosokawa, N., Hara, Y. & Mizushima, N. Generation of cell lines with tetracycline-regulated autophagy and a role for autophagy in controlling cell size. FEBS Lett. 580, 2623–2629 (2006).
Acknowledgements
We would like to thank the individuals with SENDA and their families for their participation in this study. We thank M. Shiina and K. Ogata for their helpful comments on the protein structure. This work was supported by research grants from the Ministry of Health, Labour and Welfare (H.S., N. Miyake and N. Matsumoto), the Japan Science and Technology Agency (N. Matsumoto) and the Strategic Research Program for Brain Sciences (N. Matsumoto) and by a Grant-in-Aid for Scientific Research on Innovative Areas (Transcription Cycle) from the Ministry of Education, Culture, Sports, Science and Technology of Japan (N. Matsumoto), a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (N. Matsumoto), a Grant-in-Aid for Young Scientists from the Japan Society for the Promotion of Science (H.S. and N. Miyake), the Funding Program for Next-Generation World-Leading Researchers (N. Mizushima) and a grant from the Takeda Science Foundation (N. Miyake, N. Mizushima and N. Matsumoto).
Author information
Authors and Affiliations
Contributions
H.S., N. Mizushima and N. Matsumoto designed and directed the study. H.S., T.N., K.M., N. Mizushima and N. Matsumoto wrote the manuscript. K.M., S.K., K.S., E.K.-Y., N.S., H.N., A.H., F.R., S.Y., H.A. and M.K. collected samples and provided the subjects' clinical information. H.S., H.K., K.N., Y.T., M.N. and N. Miyake performed exome sequencing and Sanger sequencing. H.S. and K.N. performed the RNA analysis. Y.K. performed the X-inactivation analysis. T.N. and N. Mizushima analyzed protein expression and autophagic activity.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Note, Supplementary Figures 1 and 2 and Supplementary Tables 1–4 (PDF 3001 kb)
Rights and permissions
About this article
Cite this article
Saitsu, H., Nishimura, T., Muramatsu, K. et al. De novo mutations in the autophagy gene WDR45 cause static encephalopathy of childhood with neurodegeneration in adulthood. Nat Genet 45, 445–449 (2013). https://doi.org/10.1038/ng.2562
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2562
This article is cited by
-
Lipid droplet accumulation in Wdr45-deficient cells caused by impairment of chaperone-mediated autophagic degradation of Fasn
Lipids in Health and Disease (2024)
-
Mechanisms of Ferritinophagy and Ferroptosis in Diseases
Molecular Neurobiology (2024)
-
Autophagy and autophagy signaling in Epilepsy: possible role of autophagy activator
Molecular Medicine (2023)
-
The HCN1 p.Ser399Pro variant causes epileptic encephalopathy with super-refractory status epilepticus
Human Genome Variation (2023)
-
Skewed X-chromosome Inactivation in Women with Idiopathic Intellectual Disability is Indicative of Pathogenic Variants
Molecular Neurobiology (2023)